Search Count: 7
 |
L122K Mutant Of Fmn-Binding Protein From Desulfovibrio Vulgaris (Miyazaki F)
Organism: Desulfovibrio vulgaris str. 'miyazaki f'
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-05-19 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
L122Y Mutant Of Fmn-Binding Protein From Desulfovibrio Vulgaris (Miyazaki F)
Organism: Desulfovibrio vulgaris str. 'miyazaki f'
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-07-19 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
L122E Mutant Of Fmn-Binding Protein From Desulfovibrio Vulgaris (Miyazaki F)
Organism: Desulfovibrio vulgaris str. 'miyazaki f'
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-07-19 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-11-16 Classification: HYDROLASE Ligands: CA, SO4 |
 |
Crystal Structure Of A New Alkaline Serine Protease (M-Protease) From Bacillus Sp. Ksm-K16
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-06-22 Classification: SERINE PROTEINASE Ligands: CA |
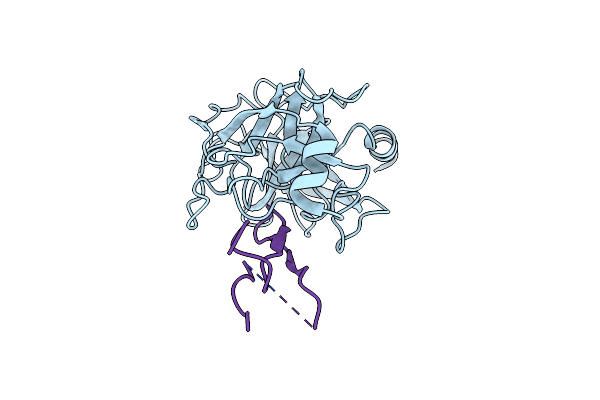 |
Structure Of The Trypsin-Binding Domain Of Bowman-Birk Type Protease Inhibitor And Its Interaction With Trypsin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1992-01-15 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
The Crystal Structure Of Bonito (Katsuo) Ferrocytochrome C At 2.3 Angstroms Resolution. Ii. Structure And Function
Organism: Katsuwonus pelamis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1976-10-06 Classification: ELECTRON TRANSPORT Ligands: HEC |

