Search Count: 98
 |
Organism: Ascaris suum
Method: SOLUTION NMR Release Date: 2022-09-21 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Ascaris suum
Method: SOLUTION NMR Release Date: 2021-11-17 Classification: ANTIMICROBIAL PROTEIN |
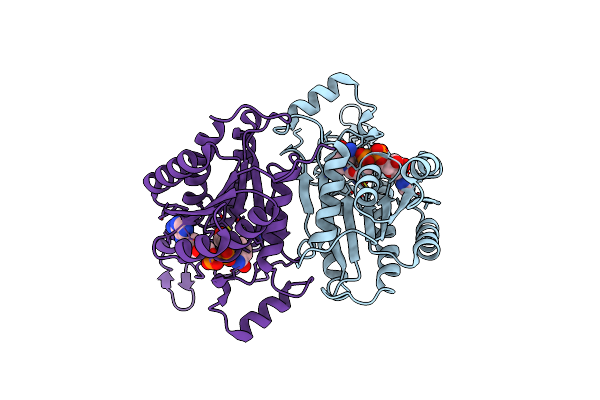 |
Structural Basis For Branched Substrate Selectivity In A Ketoreductase From Ascaris Suum
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-17 Classification: LIPID BINDING PROTEIN Ligands: VCA, EDO, TAM |
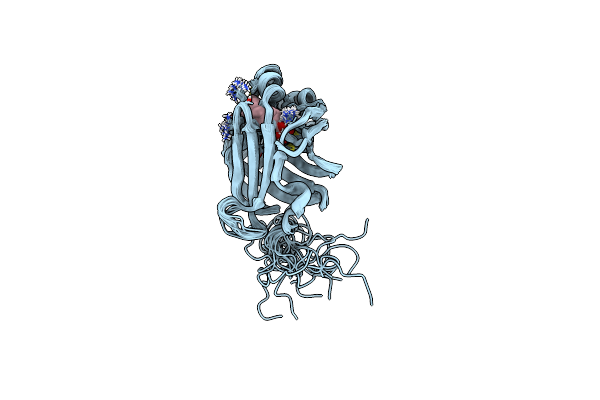 |
Solution Structure Of As-P18 Reveals That Nematode Fatty Acid Binding Proteins Exhibit Unusual Structural Features
Organism: Ascaris suum
Method: SOLUTION NMR Release Date: 2019-07-17 Classification: LIPID BINDING PROTEIN Ligands: OLA |
 |
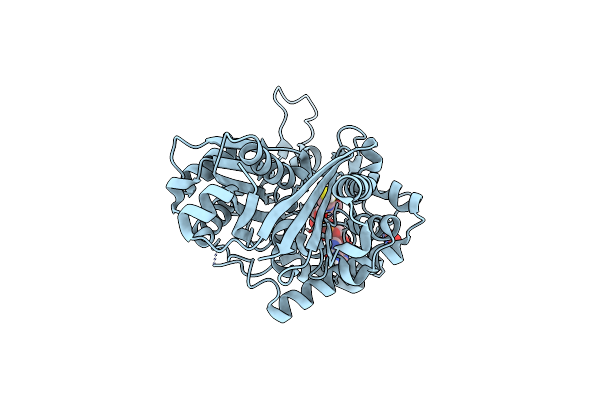
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2018-02-07 Classification: TRANSFERASE Ligands: K |
 |
Crystal Structure Of Acat5 Thiolase From Ascaris Suum In Complex With Coenzyme A
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-02-07 Classification: TRANSFERASE Ligands: ACT, COA |
 |
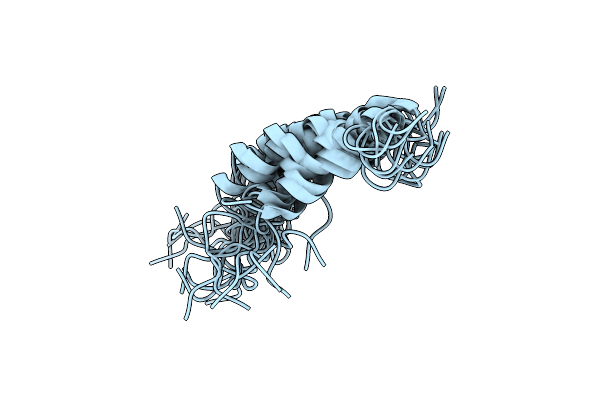
Crystal Structure Of Acat2-C91S Thiolase From Ascaris Suum In Complex With Propionyl-Coa And Nitrate
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-02-07 Classification: TRANSFERASE Ligands: 1VU, NO3 |
 |
Organism: Ascaris suum
Method: SOLUTION NMR Release Date: 2016-11-09 Classification: ANTIMICROBIAL PROTEIN |
 |
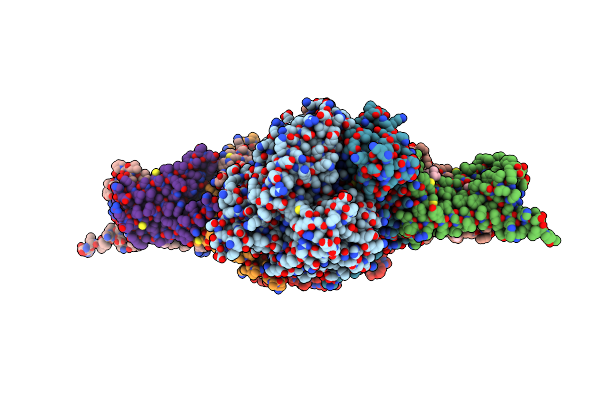
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With Ubiquinone-1
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, UQ1, EPH |
 |
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With The Specific Inhibitor Nn23
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, E23, HEM, EPH |
 |
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With N-[(2,4-Dichlorophenyl)Methyl]-2-(Trifluoromethyl)Benzamide
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, E24, HEM, EPH |
 |
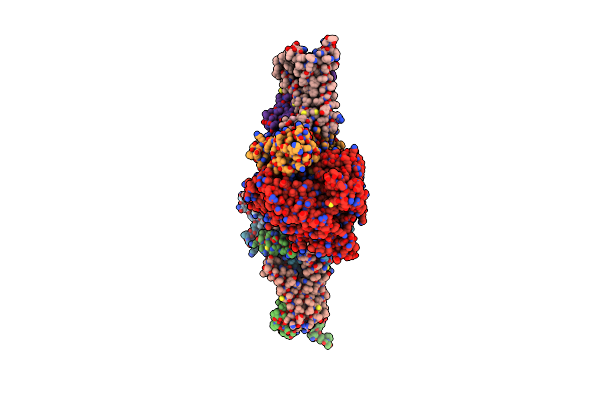
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With 2-Iodo-N-[3-(1-Methylethoxy)Phenyl]Benzamide
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, 12J, EPH |
 |
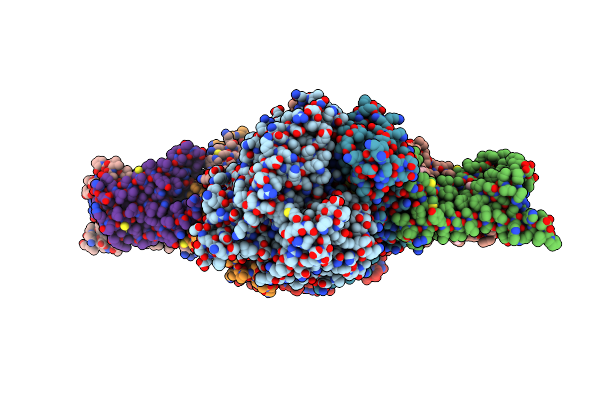
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With 2-Methyl-N-[3-(1-Methylethoxy)Phenyl]Benzamide.
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.66 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, MRN |
 |
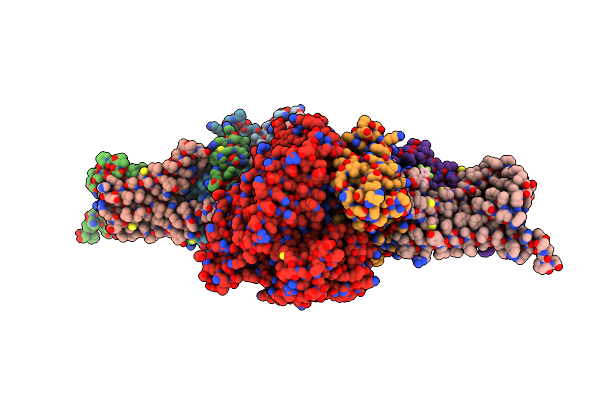
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With N-Biphenyl-3-Yl-2-(Trifluoromethyl)Benzamide
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, F6A, EPH |
 |
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With N-[3-(Pentafluorophenoxy)Phenyl]-2-(Trifluoromethyl)Benzamide
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, FD8 |
 |
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With Rhodoquinone-2
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, 4YP, EPH |
 |
Crystal Structure Of The Glutathione S-Transferase From Ascaris Lumbricoides
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-04-01 Classification: TRANSFERASE Ligands: GSH |
 |
Mitochondrial Rhodoquinol-Fumarate Reductase From The Parasitic Nematode Ascaris Suum With The Specific Inhibitor Flutolanil
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2013-04-10 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, FTN, EPH |
 |
Mitochondrial Rhodoquinol-Fumarate Reductase From The Parasitic Nematode Ascaris Suum With The Specific Inhibitor Atpenin A5
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.44 Å Release Date: 2013-04-10 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, AT5, EPH |

