Search Count: 89
 |
Organism: Pseudomonas sp.
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Discovery And Characterization Of A New Carbonyl Reductase From Rhodotorula Toluroides Reducing Fluoroketones, And X-Ray Analysis Of The Variant By Rational Engineering
Organism: Rhodotorula toruloides
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: MG |
 |
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: HBX, CL, SO4, GOL |
 |
Hydroxynitrile Lyase From The Millipede, Oxidus Gracilis Bound With (R)-(+)-Alpha-Hydroxybenzene-Acetonitrile
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: MXN, SO4 |
 |
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: CNH, CL, SO4 |
 |
Hydroxynitrile Lyase From The Millipede, Oxidus Gracilis Complexed With (R)-2-Chloromandelonitrile
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: IJ5, GOL, SO4 |
 |
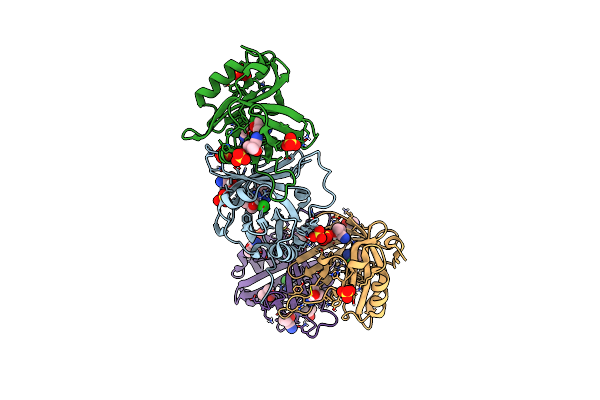
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-17 Classification: LYASE Ligands: SO4, CL |
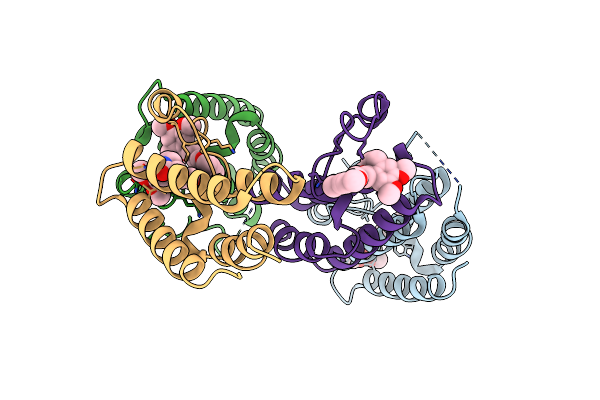 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-08-03 Classification: TRANSCRIPTION Ligands: 4JI, CL |
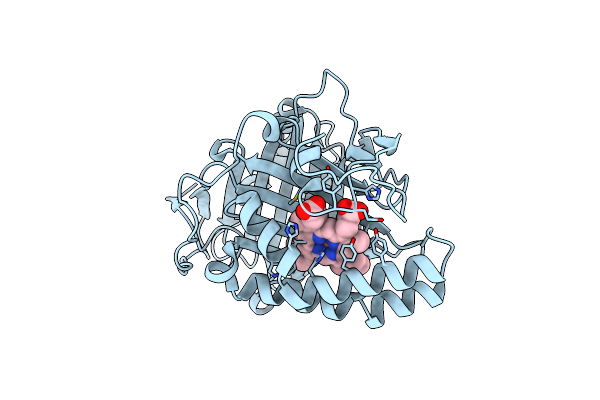 |
Organism: Bacillus sp. (strain oxb-1)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-04-27 Classification: LYASE Ligands: HEM |
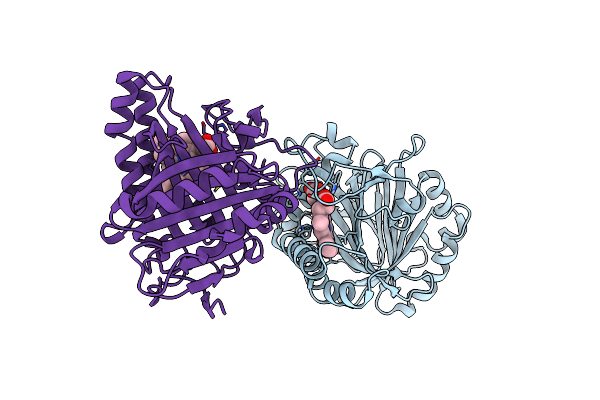 |
Organism: Bacillus sp. (strain oxb-1)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-04-27 Classification: LYASE Ligands: HEM |
 |
Crystal Structure Of Oxdb E85A In Complex With Z-2- (3-Bromophenyl) Propanal Oxime
Organism: Bacillus sp. (strain oxb-1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-27 Classification: LYASE Ligands: HEM, 0WQ |
 |
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, GOL, MG, EDO, BTB |
 |
Crystal Structure Of Hydroxynitrile Lyase From Linum Usitatissimum Complexed With Acetone Cyanohydrin
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, CNH, MG, EDO, GOL, BTB, ACN |
 |
Crystal Structure Of Hydroxynitrile Lyase From Linum Usitatissium Complexed With (R)-2-Hydroxy-2-Methylbutanenitrile
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, MG, 5Z9, EDO, GOL, BTB |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE Ligands: NAD, THR, PE4 |
 |
Hydroxynitrile Lyase From Parafontaria Laminate Complexed With Benzaldehyde Prepared By Cocrystallization
Organism: Parafontaria laminata
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2021-04-07 Classification: LYASE Ligands: HBX, SCN, EDO |
 |
Organism: Parafontaria falcifera
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2021-03-24 Classification: LYASE Ligands: GOL |
 |
Hydroxynitrile Lyase From Parafonteria Laminate Complexed With Benzaldehyde
Organism: Parafontaria falcifera
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2021-03-24 Classification: LYASE Ligands: HBX, GOL |

