Search Count: 110
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-01-17 Classification: SIGNALING PROTEIN Ligands: PEG, MG, BEF |
 |
Crystal Structure Of Glucokinase (Hexokinase 4) Complexed With Ligand Diethyl ({2-[3-(4-Methanesulfonylpheno Xy)-5-{[(2S)-1-Methoxypropan-2-Yl]Oxy}Benzamido]-1,3-Thiaz Ol-4-Yl}Methyl)Phosphonate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-03-02 Classification: TRANSFERASE Ligands: GLC, NA, G2T, EDO |
 |
Crystal Structure Of Glucokinase (Hexokinase 4) Complexed With Ligand Aka Diethyl {[3-(3-{[5-(Azetidine-1-Carbon Yl)Pyrazin-2-Yl]Oxy}-5-(Propan-2-Yloxy)Benzamido)-1H- Pyrazol-1-Yl]Methyl}Phosphonate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-03-02 Classification: TRANSFERASE Ligands: G1S, GLC |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-07 Classification: SIGNALING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-04-07 Classification: SIGNALING PROTEIN Ligands: MG, BEF |
 |
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, GOL |
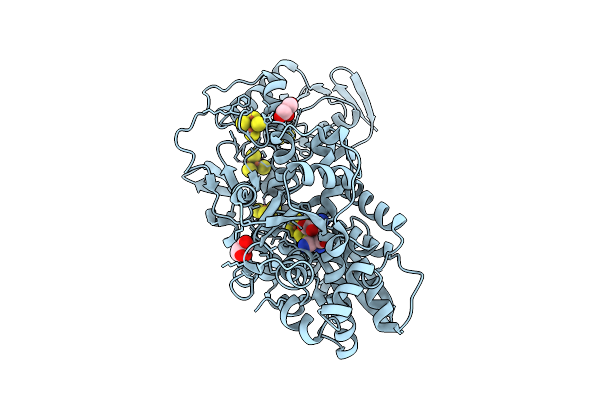 |
Crystal Structure Of [Fefe]-Hydrogenase In The Presence Of 7 Mm Sodium Dithionite
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, GOL |
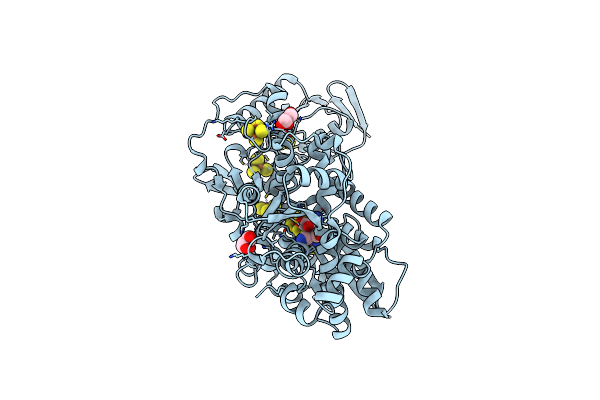 |
Crystal Structure Of [Fefe]-Hydrogenase I (Cpi) Solved With Single Pulse Free Electron Laser Data
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2019-05-08 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-05-08 Classification: DE NOVO PROTEIN |
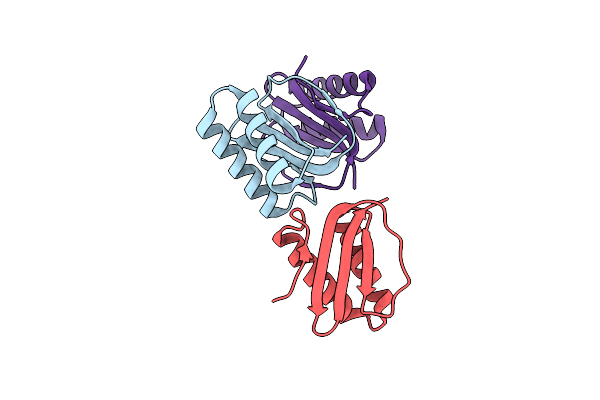 |
1.65 Angstrom Structure Of The Dynein Light Chain 1 From Plasmodium Falciparum
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-08-16 Classification: TRANSPORT PROTEIN |
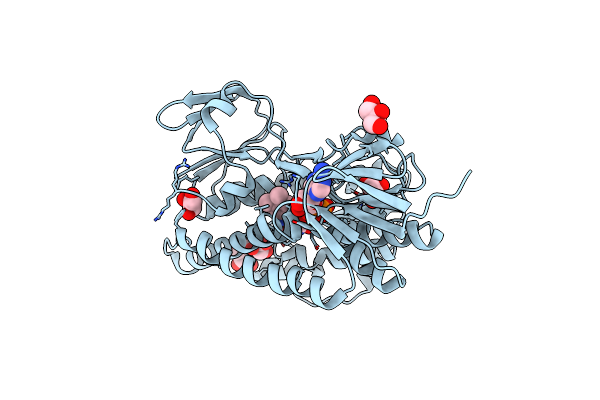 |
Organism: Metallosphaera sedula (strain atcc 51363 / dsm 5348)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2015-09-16 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
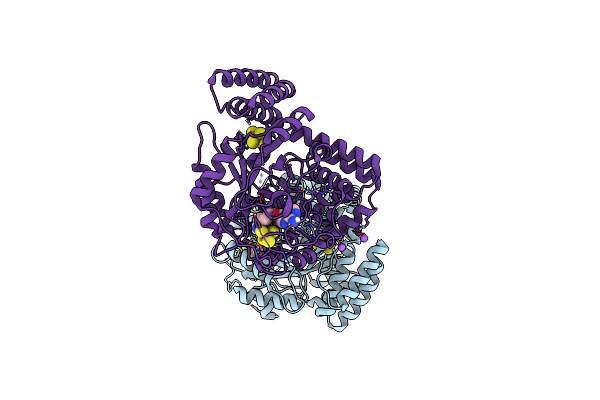 |
Organism: Thermoanaerobacter italicus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2015-02-04 Classification: LYASE Ligands: SF4, MET, FE, ALA, H2S, NA, SAM |
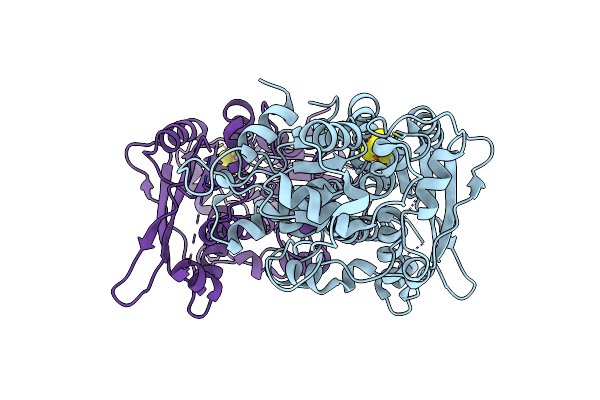 |
[Fefe]-Hydrogenase Oxygen Inactivation Is Initiated By The Modification And Degradation Of The H Cluster 2Fe Subcluster
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2015-01-28 Classification: OXIDOREDUCTASE Ligands: SF4, CL, ARS |
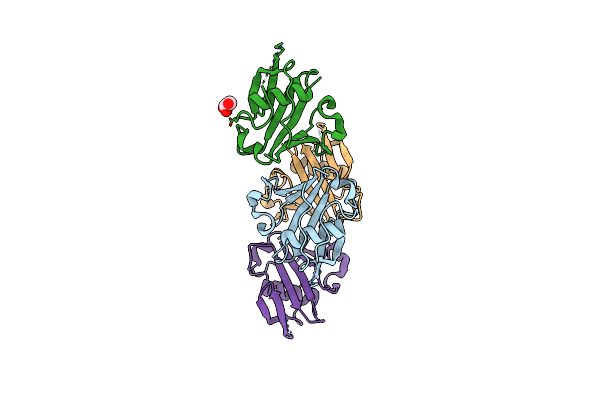 |
Organism: Plasmodium yoelii yoelii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-12-25 Classification: PROTEIN BINDING Ligands: GOL |
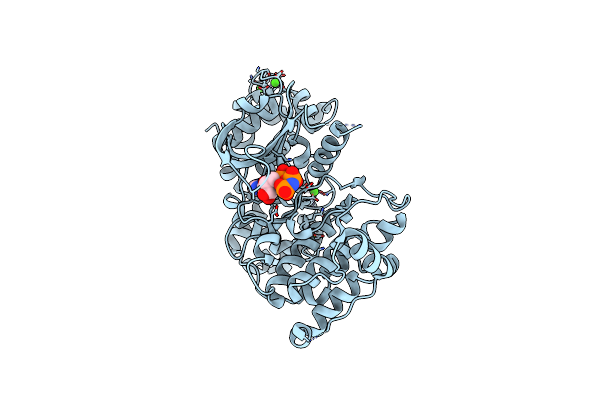 |
Crystal Structure Of A Gly128Met Mutant Of The Toxoplasma Cdpk, Tgme49_101440
Organism: Toxoplasma gondii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-12-25 Classification: TRANSFERASE Ligands: CA, ANP |
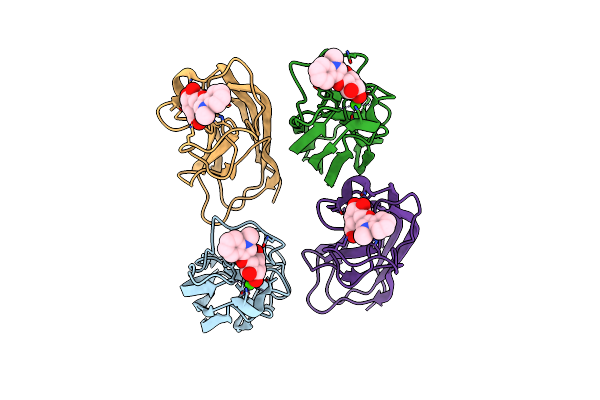 |
Crystal Structure Of Pseudomonas Aeruginosa Lectin Leca Complexed With 1-Methyl-3-Indolyl-B-D-Galactopyranoside At 1.45 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2013-10-30 Classification: SUGAR BINDING PROTEIN Ligands: CA, MHD, GAL |
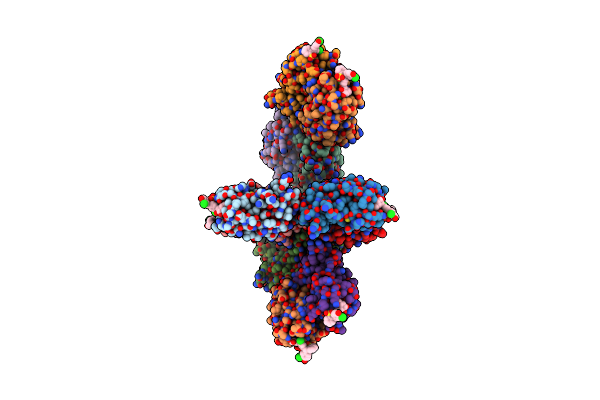 |
Crystal Structure Of Pseudomonas Aeruginosa Lectin Leca Complexed With Chlorophenol Red-B-D-Galactopyranoside At 2.86 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2013-10-30 Classification: SUGAR BINDING PROTEIN Ligands: CA, LRD, GAL |
 |
Crystal Structure Of Pseudomonas Aeruginosa Lectin Leca Complexed With Resorufin-B-D-Galactopyranoside At 1.76 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2013-10-30 Classification: SUGAR BINDING PROTEIN Ligands: CA, 04G, GAL |
 |
Organism: Plasmodium yoelii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-10-19 Classification: STRUCTURAL GENOMICS,UNKNOWN FUNCTION Ligands: SO4, GOL, EDO |

