Search Count: 41
 |
Organism: Escherichia phage t5
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: MG, CL, EDO, B3P, IOD |
 |
Organism: Escherichia phage t5
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: EDO, MG, CL |
 |
Organism: Escherichia phage t5, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: MG, K, GOL |
 |
Crystal Structure Bacteriohage T5 D15 Flap Endonuclease (D155K) Pseudo-Enzyme-Product Complex With Dna And Metal Ions
Organism: Escherichia phage t5, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: CA, NA, K, GOL |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-09-18 Classification: TOXIN Ligands: EDO, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-07-10 Classification: HYDROLASE Ligands: K |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-07-10 Classification: HYDROLASE Ligands: K |
 |
Structure Of E. Coli Exoix In Complex With A Fragment Of The Flap1 Dna Oligonucleotide, Potassium And Magnesium
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-07-10 Classification: HYDROLASE/DNA Ligands: K, MG, PIV, PO4 |
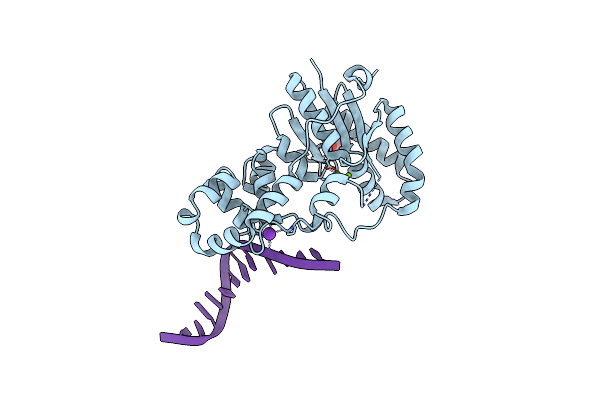 |
Structure Of E. Coli Exoix In Complex With The Palindromic 5Ov4 Dna Oligonucleotide, Di-Magnesium And Potassium
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2013-07-10 Classification: HYDROLASE/DNA Ligands: K, MG, ACT |
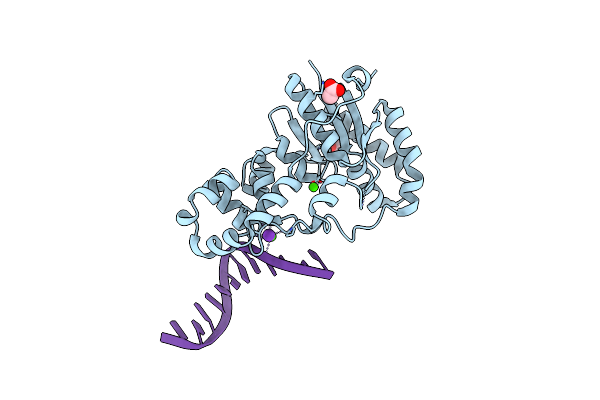 |
Structure Of E. Coli Exoix In Complex With The Palindromic 5Ov4 Dna Oligonucleotide, Potassium And Calcium
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2013-07-10 Classification: HYDROLASE Ligands: K, CA, ACT |
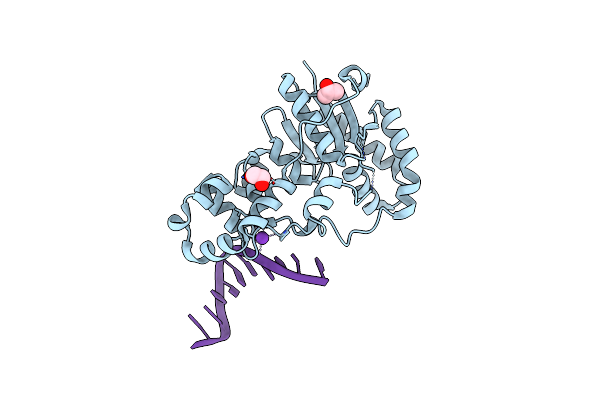 |
Structure Of E. Coli Exoix In Complex With The Palindromic 5Ov6 Oligonucleotide And Potassium
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-07-10 Classification: HYDROLASE/DNA Ligands: K, IPA, EDO |
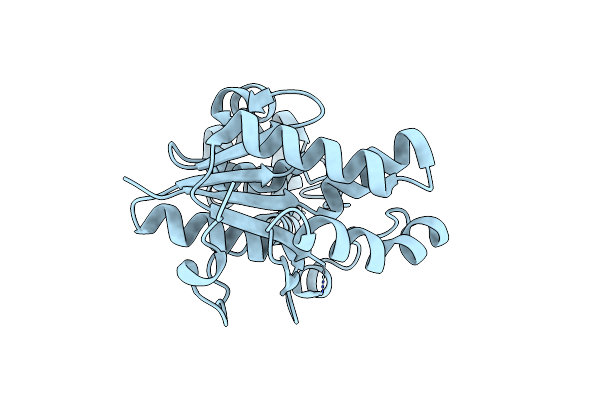 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2013-07-10 Classification: HYDROLASE |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-03-14 Classification: IMMUNE SYSTEM Ligands: CYS, EDO, ACT, NA, NAG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2012-03-14 Classification: IMMUNE SYSTEM Ligands: ACT, SO4 |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2011-11-30 Classification: UNKNOWN FUNCTION Ligands: BR |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2011-11-30 Classification: UNKNOWN FUNCTION |
 |
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2005-08-30 Classification: DNA BINDING PROTEIN |
 |
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2005-08-30 Classification: DNA BINDING PROTEIN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-05-25 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2004-05-25 Classification: HYDROLASE Ligands: GDP |

