Search Count: 38
 |
Organism: Streptomyces avidinii, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: PROTEIN BINDING Ligands: GOL |
 |
Crystal Structure Of Complement1.4 (Cent1.4), An Engineered Photoenzyme For Selective [2+2]-Cycloadditions
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, PEG, PGE |
 |
Engineered Lmrr With V15 Replaced By Unnatural Amino Acid 4-Amino-L-Phenyl-Cysteine
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: TRANSCRIPTION Ligands: EDO |
 |
Crystal Structure Of Lmrr Variant V15Ay With Val15 Replaced By 3-Aminotyrosine
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Lmrr Variant V15Ay-Rnyw With Val15 Replaced By 3-Aminotyrosine And Evolved As Friedel-Crafts Alkylase
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA BINDING PROTEIN Ligands: NO3 |
 |
Carbonic Anhydrase Ii Variant With Bound Iron Complex In Space Group R3 (Arpase)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-03-05 Classification: OXIDOREDUCTASE Ligands: A1H9O, ZN, EDO |
 |
Crystal Structure Of Artificial Enzyme Lmrr_Paf Variant Rmh In Crystal Form 1
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Artificial Enzyme Lmrr_Paf Variant Rmh In Crystal Form 2
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Ramr With Tyr59 Replaced With Para-Boronophenylalanine (Boronate Form)
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-12-25 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Heme-Oxygenase From Corynebacterium Diphtheriae Complexed With Cobalt-Porphyrine (Humo-Co(Iii))
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: COH, GOL, EDO, CIT, CL |
 |
Crystal Structure Of Heme-Oxygenase From Corynebacterium Diphtheriae Complexed With Cobalt-Porphyrine (Humo-Co(Iii)) Flash-Cooled Under Co2 Pressure
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: COH, EDO, CL, CO2 |
 |
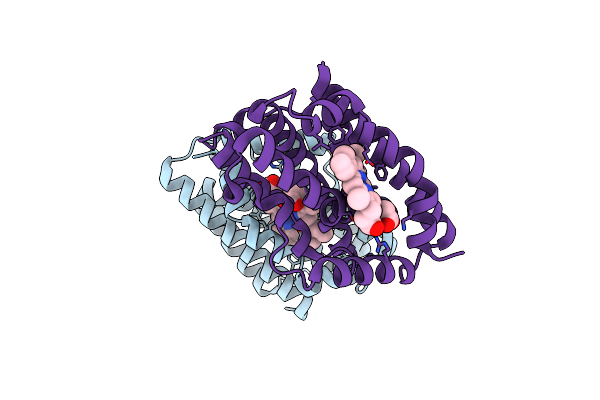
Crystal Structure Of Heme-Oxygenase Mutant G139A From Corynebacterium Diphtheriae Complexed With Cobalt-Porphyrine (Humo-Co(Iii))
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: COH, CL |
 |
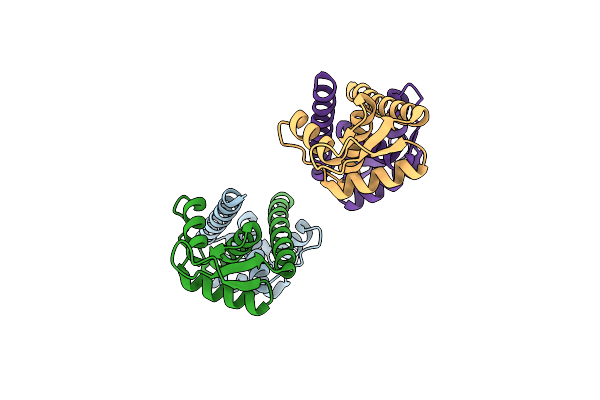
Crystal Structure Of Heme-Oxygenase Mutant H20C From Corynebacterium Diphtheriae Complexed With Cobalt-Porphyrine (Humo-Co(Iii))
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: COH |
 |
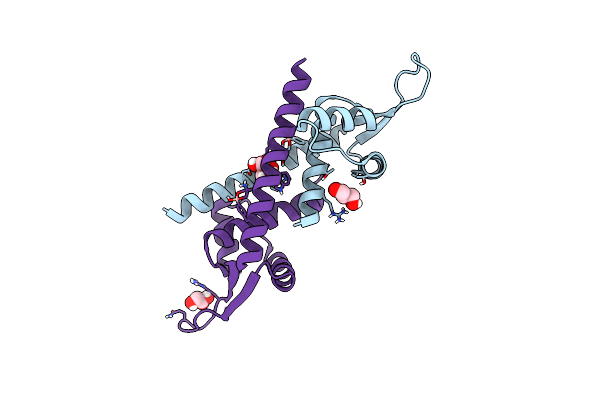
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-05-01 Classification: TRANSCRIPTION |
 |
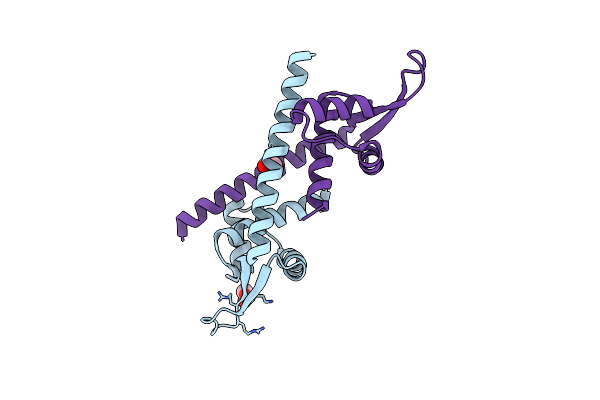
Engineered Lmrr Carrying A Cyclic Boronate Ester Formed Between Tris And P-Boronophenylalanine At Position 89
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-05-01 Classification: TRANSCRIPTION Ligands: GOL |
 |
Crystal Structure Of Lmrr With V15 Replaced By Unnatural Amino Acid 4-Amino-L-Phenyl-Cysteine
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2024-04-24 Classification: DNA BINDING PROTEIN Ligands: EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-01-10 Classification: LYASE Ligands: PO4, MES |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-01-10 Classification: LYASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2024-01-10 Classification: LYASE |

