Search Count: 1,673
All
Selected
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured By Mix-And-Inject Serial Crystallography At 100-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: PEA, NA, CU |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured By Mix-And-Inject Serial Crystallography At 200-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: PEA, NA, CU |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured By Mix-And-Inject Serial Crystallography At 300-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured By Mix-And-Inject Serial Crystallography At 400-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: PEA, NA, CU |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured With Short-A-Axis Diffraction Data By Mix-And-Inject Serial Crystallography At 22-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: CU, NA |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured With Long-A-Axis Diffraction Data By Mix-And-Inject Serial Crystallography At 22-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: CU, NA |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured With Short-A-Axis Diffraction Data By Mix-And-Inject Serial Crystallography At 25-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured With Long-A-Axis Diffraction Data By Mix-And-Inject Serial Crystallography At 25-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: CU, NA |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured With Short-A-Axis Diffraction Data By Mix-And-Inject Serial Crystallography At 50-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured With Long-A-Axis Diffraction Data By Mix-And-Inject Serial Crystallography At 50-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: CU, NA |
 |
Oxidative Form Of Copper Amine Oxidase From Arthrobacter Globiformis Determined At Ph 9.0 By Single-Flow Serial Crystallography
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: CU, NA |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured By Mix-And-Inject Serial Crystallography At 500-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: CU, NA, PEA |
 |
Reductive-Half Reaction Intermediate Of Copper Amine Oxidase From Arthrobacter Globiformis Captured By Mix-And-Inject Serial Crystallography At 1000-Ms Time Delay
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: CU, NA, PEA |
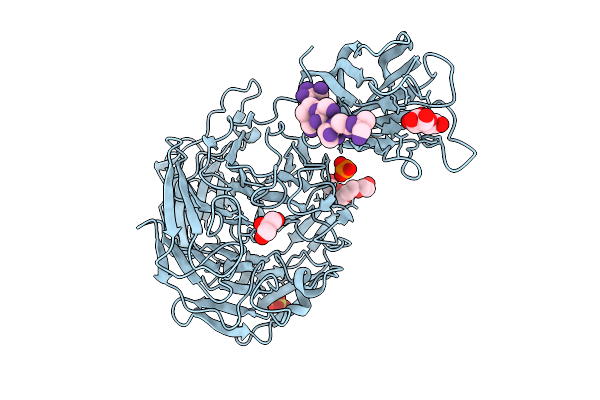 |
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: GAL, PG4, GOL, PO4, CA, SO4 |
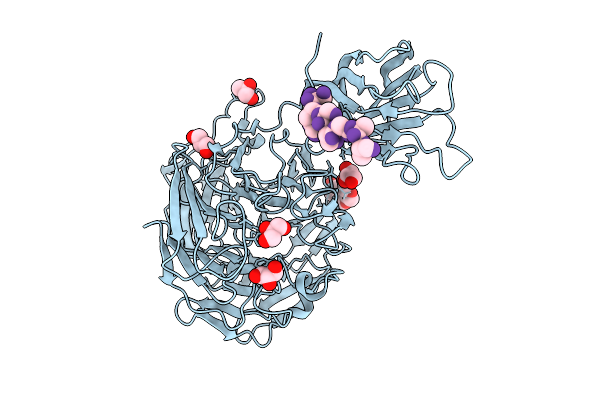 |
Crystal Structure Of A Galactose Oxidase From Pseudoarthrobacter Siccitolerans
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: GOL, PG4, CA, CL, BR |
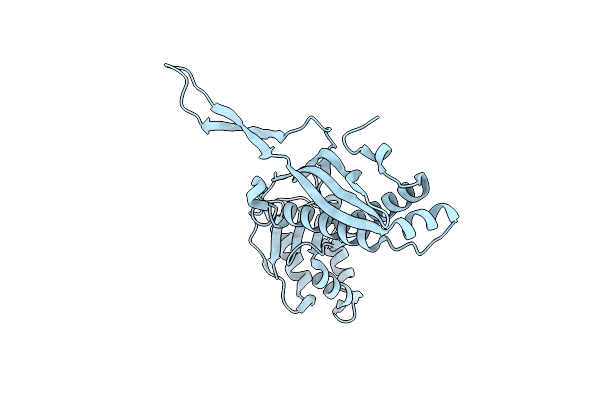 |
Organism: Arthrobacter sp.
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-05-21 Classification: CYTOSOLIC PROTEIN |
 |
Crystal Structure Of L-Ribulose 3-Epimerase From Arthrobacter Globiformis M30
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-02-12 Classification: ISOMERASE Ligands: MG, PEG |
 |
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-02-12 Classification: ISOMERASE Ligands: PSJ, FUD, MG, NA, PEG |
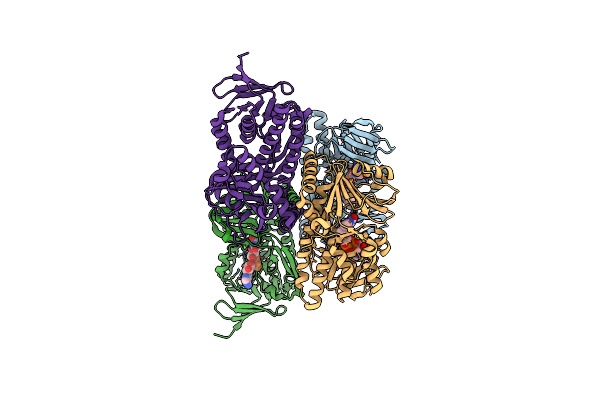 |
Organism: Arthrobacter sp. pamc25564
Method: X-RAY DIFFRACTION Resolution:3.29 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Gh9L Inulin Fructotransferases (Iftase) In Compex With Fruetosyl Nystose (Gf4)
Organism: Paenarthrobacter aurescens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-09-04 Classification: CARBOHYDRATE |

