Search Count: 31
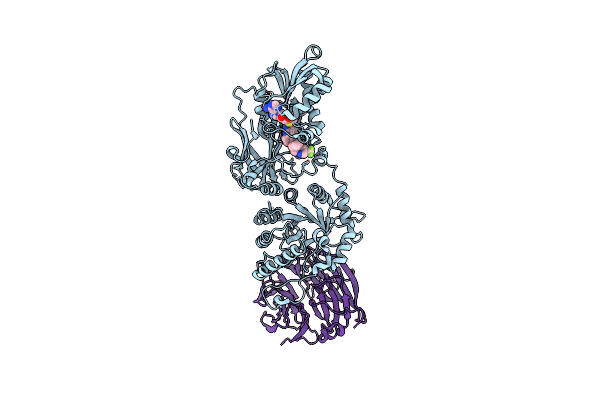 |
Discovery Of Mta-Cooperative Prmt5 Inhibitors From Co-Factor Directed Dna-Encoded Library Screens
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-05-07 Classification: TRANSFERASE/INHIBITOR Ligands: A1B6Y, MTA |
 |
Solution-State Nmr Structural Ensemble Of Human Tsg101 Uev In Complex With K63-Linked Diubiquitin
|
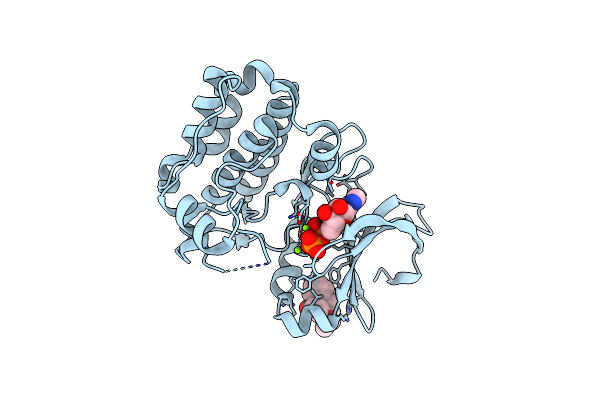 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2019-05-08 Classification: TRANSFERASE Ligands: ADP, MG, JSB |
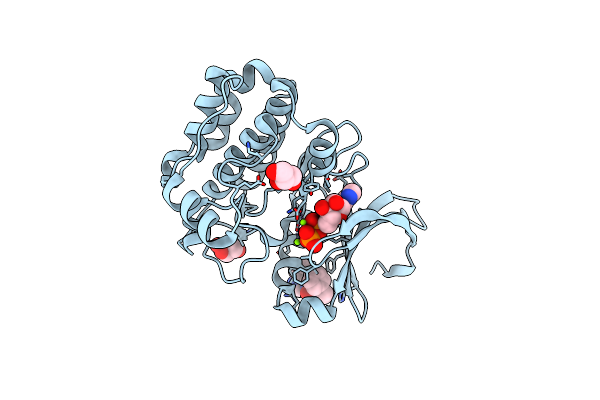 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-05-08 Classification: TRANSFERASE Ligands: ADP, GOL, MG, CL, JRT |
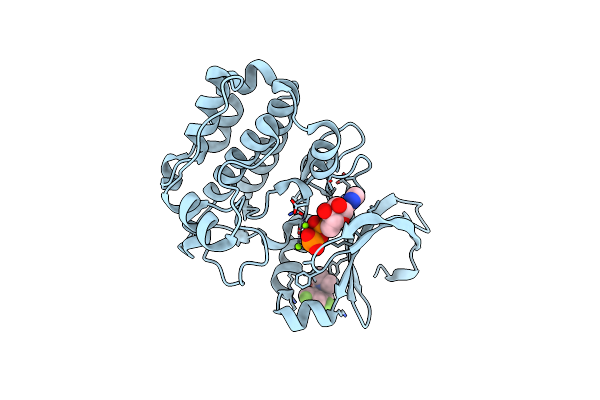 |
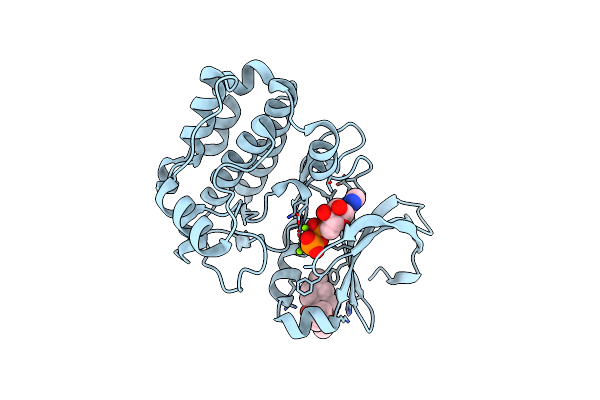
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-05-08 Classification: TRANSFERASE Ligands: ADP, MG, CL, JSN |
 |
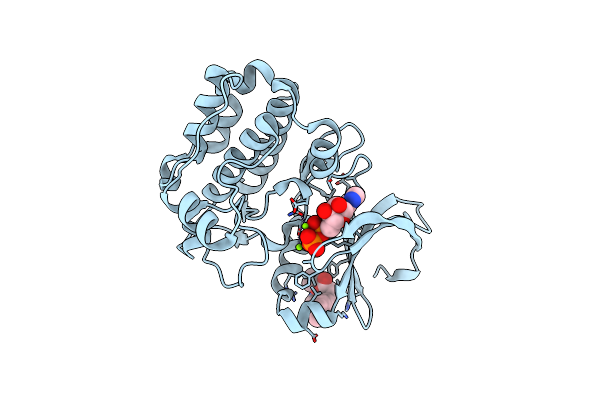
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2019-05-08 Classification: TRANSFERASE Ligands: ADP, MG, CL, JRW |
 |
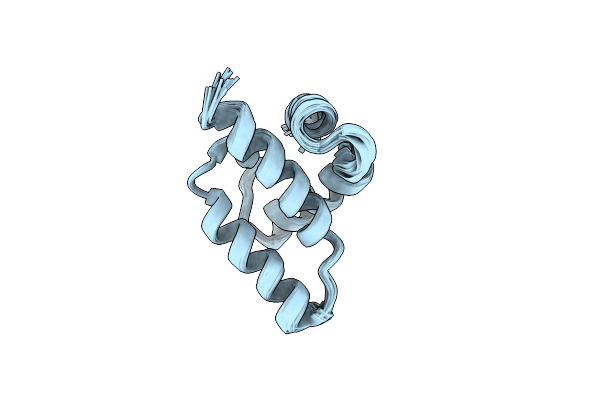
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2019-05-01 Classification: TRANSFERASE Ligands: ADP, MG, CL, JRQ |
 |
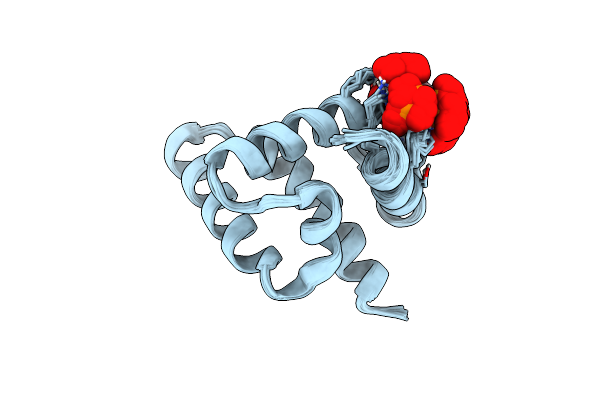
Nmr Structure Of The Rous Sarcoma Virus Matrix Protein (M-Domain) In The Presence Of Myo-Inositol Hexakisphosphate
Organism: Rous sarcoma virus
Method: SOLUTION NMR Release Date: 2018-11-07 Classification: VIRAL PROTEIN |
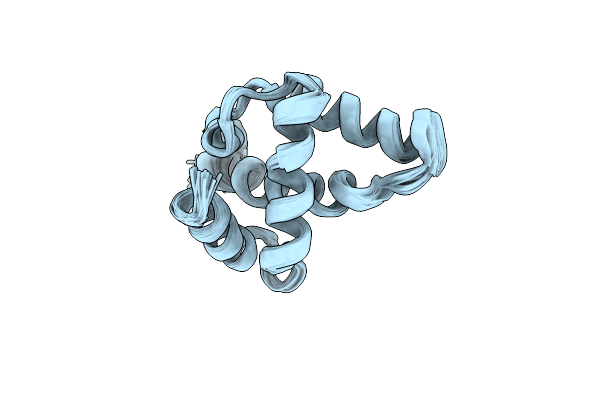 |
Organism: Rous sarcoma virus (strain prague c)
Method: SOLUTION NMR Release Date: 2018-10-24 Classification: VIRAL PROTEIN |
 |
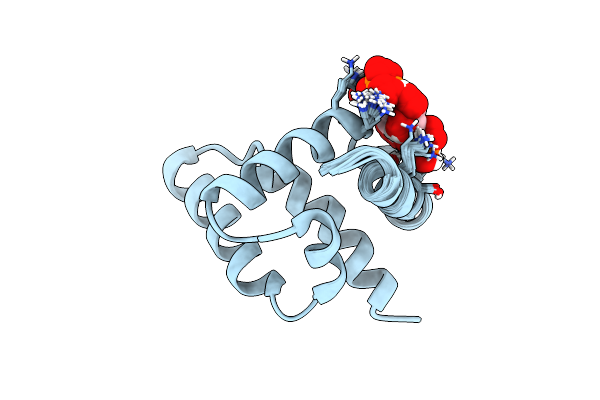
Haddock Structure Of The Rous Sarcoma Virus Matrix Protein (M-Domain) In Complex With Myo-Inositol Hexakisphosphate
Organism: Rous sarcoma virus
Method: SOLUTION NMR Release Date: 2018-10-24 Classification: VIRAL PROTEIN Ligands: IHP |
 |
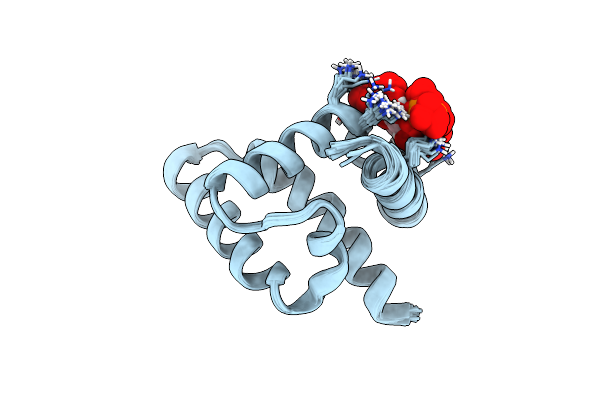
Haddock Structure Of The Rous Sarcoma Virus Matrix Protein (M-Domain) In Complex With Inositol 1,4,5-Trisphosphate
Organism: Rous sarcoma virus
Method: SOLUTION NMR Release Date: 2018-10-24 Classification: VIRAL PROTEIN Ligands: I3P |
 |
Haddock Structure Of The Rous Sarcoma Virus Matrix Protein (M-Domain) In Complex With Inositol 1,3,5-Trisphosphate
Organism: Rous sarcoma virus
Method: SOLUTION NMR Release Date: 2018-10-24 Classification: VIRAL PROTEIN Ligands: FGV |
 |
Solution-State Nmr Structural Ensemble Of Human Tsg101 Uev In Complex With Tenatoprazole
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2017-11-15 Classification: cell cycle/inhibitor Ligands: 4N1 |
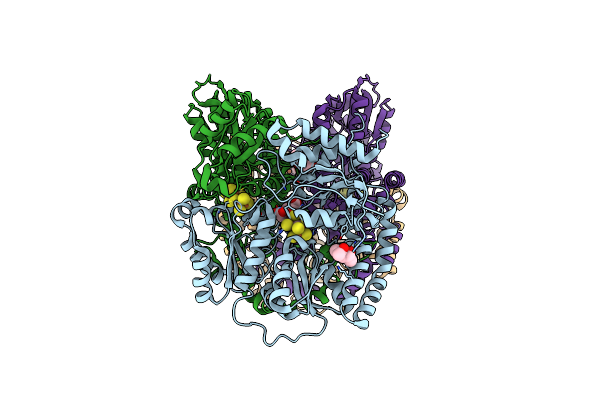 |
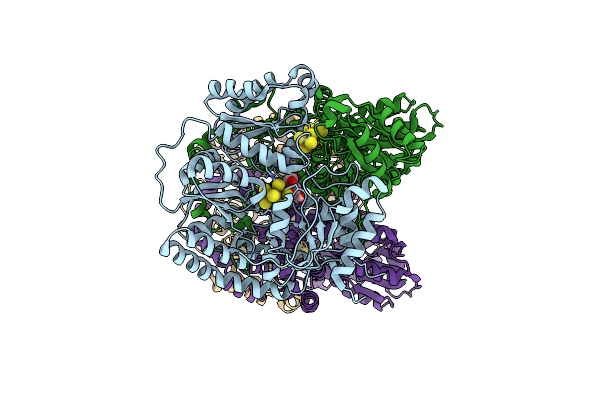
Nitrogenase Mofep From Gluconacetobacter Diazotrophicus In Dithionite Reduced State
Organism: Gluconacetobacter diazotrophicus (strain atcc 49037 / dsm 5601 / pal5)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE Ligands: HCA, ICS, MRD, CLF, FE, MPD |
 |
Organism: Gluconacetobacter diazotrophicus (strain atcc 49037 / dsm 5601 / pal5)
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-09-14 Classification: OXIDOREDUCTASE Ligands: HCA, ICS, CLF, FE |
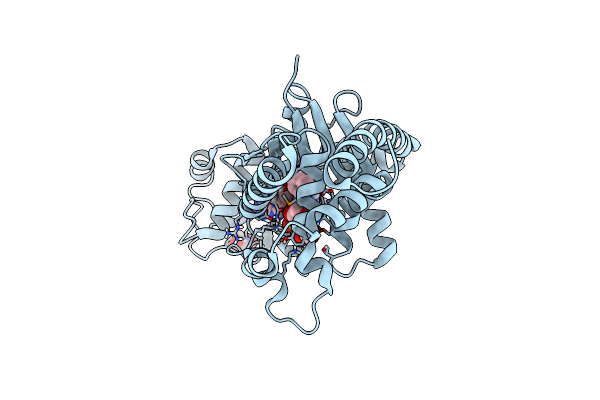 |
Crystal Structure Analysis Of Tryptophanyl-Trna Synthetase From Bacillus Stearothermophilus In Complex With Indolmycin And Mg*Atp
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-11-18 Classification: LIGASE/LIGASE INHIBITOR Ligands: ATP, MG, GOL, 5BX |
 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-09-23 Classification: OXIDOREDUCTASE Ligands: HCA, ICS, CLF, CA |
 |
Independent Saturation Of Three Trprs Subsites Generates A Partially-Assembled State Similar To Those Observed In Molecular Simulations
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2009-02-03 Classification: TRANSLATION Ligands: TRP, PO4, AMP |
 |
Crystal Structure Analysis Of B. Stearothermophilus Tryptophanyl-Trna Synthetase Complexed With Tryptophan, Amp, And Inorganic Phosphate
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-02-03 Classification: TRANSLATION Ligands: TRP, PO4, AMP |
 |
Crystal Structure Of Tryptophanyl-Trna Synthetase Complexed With Atp And Tryptophanamide In A Pre-Transition State Conformation
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2003-01-07 Classification: LIGASE |

