Search Count: 63
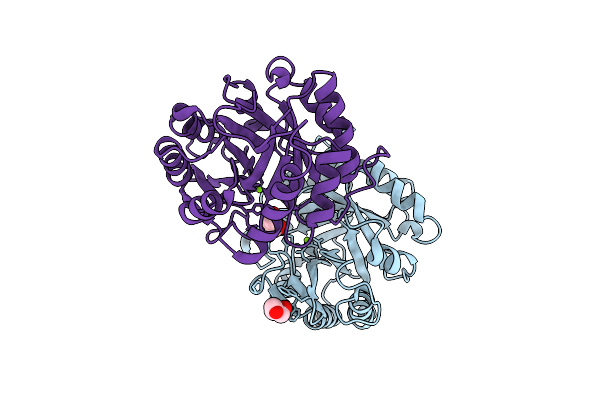 |
Crystal Structure Of Phospholipase D (Pld) From Arcanobacterium Haemolyticum
Organism: Arcanobacterium haemolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: HYDROLASE Ligands: GOL, MG |
 |
Crystal Structure Of Mutant Exfoliative Toxin C (Exhc) From Mammaliicoccus Sciuri
Organism: Mammaliicoccus sciuri
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-11-08 Classification: TOXIN |
 |
Crystal Structure Of Native Exfoliative Toxin C (Exhc) From Mammaliicoccus Sciuri
Organism: Mammaliicoccus sciuri
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2023-11-08 Classification: TOXIN |
 |
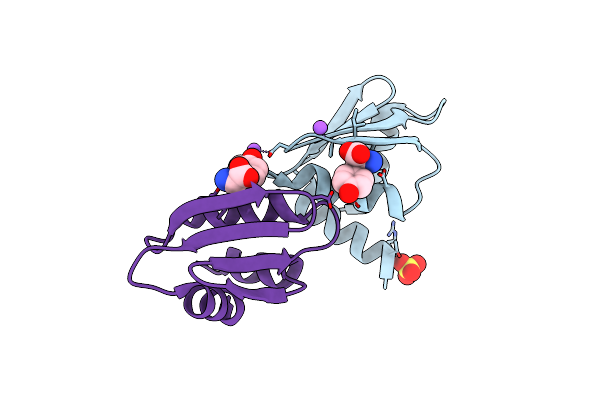
New Insights Into The P186 Flip And Oligomeric State Of Staphylococcus Aureus Exfoliative Toxin E: Implications For The Exfoliative Mechanism
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2022-11-02 Classification: TOXIN |
 |
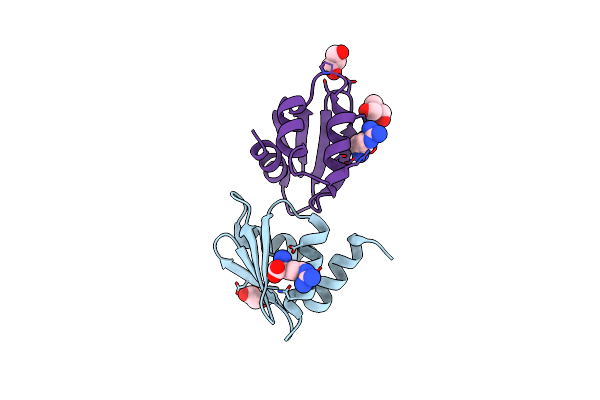
Crystal Structure Of Wild-Type Arginine Repressor From The Pathogenic Bacterium Corynebacterium Pseudotuberculosis Bound To Tyrosine
Organism: Corynebacterium pseudotuberculosis (strain c231)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2020-04-22 Classification: DNA BINDING PROTEIN Ligands: TYR, SO4, NA |
 |
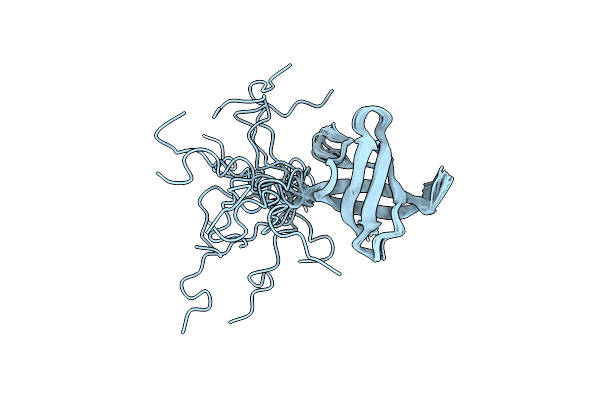
Crystal Structure Of Arginine Repressor P115Q Mutant From The Pathogenic Bacterium Corynebacterium Pseudotuberculosis Bound To Arginine
Organism: Corynebacterium pseudotuberculosis (strain c231)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-04-22 Classification: DNA BINDING PROTEIN Ligands: ARG, ACT, GOL, TRS |
 |
Nmr Structure Of Cold Shock Protein A From Corynebacterium Pseudotuberculosis
Organism: Corynebacterium pseudotuberculosis
Method: SOLUTION NMR Release Date: 2017-07-19 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of The Arginine Repressor From The Pathogenic Bacterium Corynebacterium Pseudotuberculosis
Organism: Corynebacterium pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-08-31 Classification: DNA BINDING PROTEIN Ligands: TYR, SO4 |
 |
Molecular Insights Into The Specificity Of Exfoliative Toxins From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2016-04-27 Classification: TOXIN |
 |
Crystal Structure, Maturation And Flocculating Properties Of A 2S Albumin From Moringa Oleifera Seeds
Organism: Moringa oleifera
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2015-11-11 Classification: PLANT PROTEIN Ligands: EDO, ACT |
 |
Structural Insights Into Substrate Binding Of Brown Spider Venom Class Ii Phospholipases D
Organism: Loxosceles intermedia
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-06-03 Classification: HYDROLASE Ligands: MG, PEG, IPD, GOL, OCA, SHV, DKA, PLM, TDA |
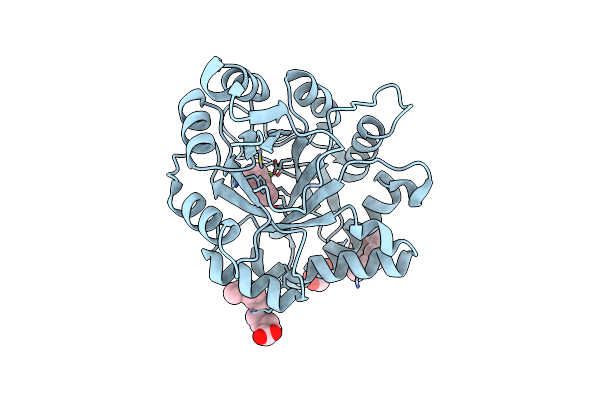 |
Structural Insights Into Substrate Binding Of Brown Spider Venom Class Ii Phospholipases D
Organism: Loxosceles intermedia
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2015-06-03 Classification: HYDROLASE Ligands: MG, PLM, GOL, TDA, OCA |
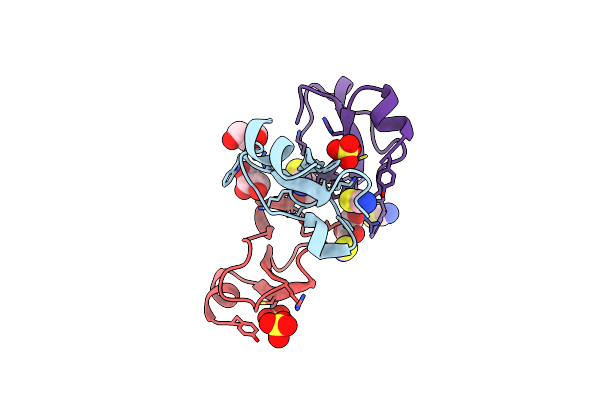 |
X-Ray Structure Of Crotamine, A Cell-Penetrating Peptide From The Brazilian Snake Crotalus Durissus Terrificus
Organism: Crotalus durissus terrificus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-09-04 Classification: TOXIN Ligands: SCN, GOL, SO4 |
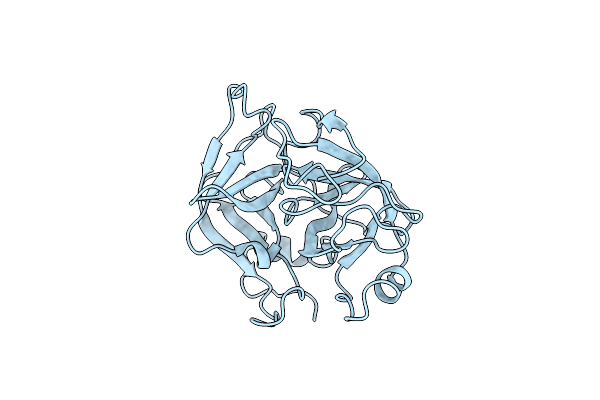 |
Organism: Bothrops jararacussu
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-12-12 Classification: HYDROLASE |
 |
Organism: Bothrops brazili
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-06-13 Classification: HYDROLASE Ligands: PG4 |
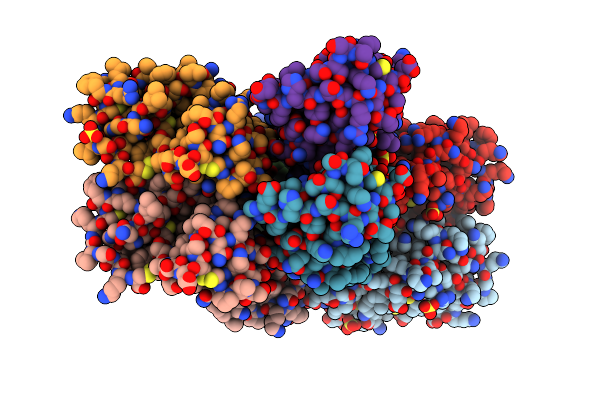 |
Crystal Structure Of A Dimeric Myotoxic Component Of The Vipera Ammodytes Meridionalis Venom Reveals Determinants Of Myotoxicity And Membrane Damaging Activity
Organism: Vipera ammodytes meridionalis
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2012-04-25 Classification: TOXIN Ligands: SO4 |
 |
Organism: Bothrops jararacussu
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2012-04-25 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Notechis scutatus scutatus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-03-28 Classification: HYDROLASE Ligands: SO4, MES |
 |
Crystal Structure Of Loxosceles Intermedia Phospholipase D Isoform 1 H12A Mutant
Organism: Loxosceles intermedia
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-08-24 Classification: HYDROLASE Ligands: MG, EDO, PEG, PGE |
 |
Crystal Structure Of A Class Ii Phospholipase D From Loxosceles Intermedia Venom
Organism: Loxosceles intermedia
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2011-06-29 Classification: HYDROLASE Ligands: A1BTZ, MG, PGE, PEG, EDO |

