Search Count: 17
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LIPID TRANSPORT Ligands: P5S |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LIPID TRANSPORT Ligands: P5S |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN |
 |
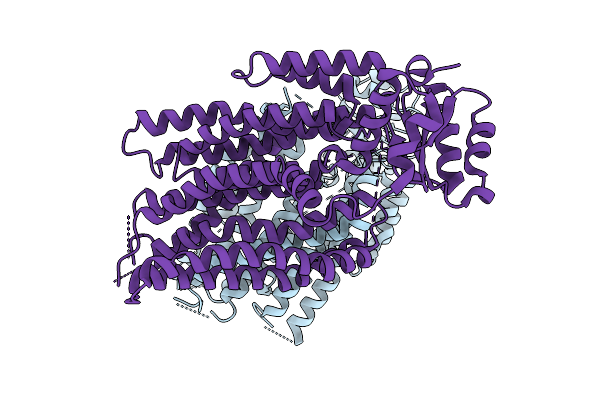
Structure Of The Transmembrane Domain Of Full-Length Ist2 At Low Resolution
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN |
 |
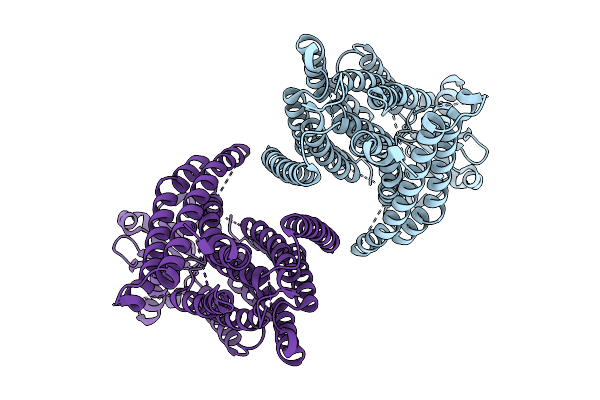
Structure Of The Transmembrane Domain Of Ist2 In Lipid Nanodiscs (Wide State)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN |
 |
Structure Of The Transmembrane Domain Of Ist2 In Lipid Nanodiscs (Narrow State)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN |
 |
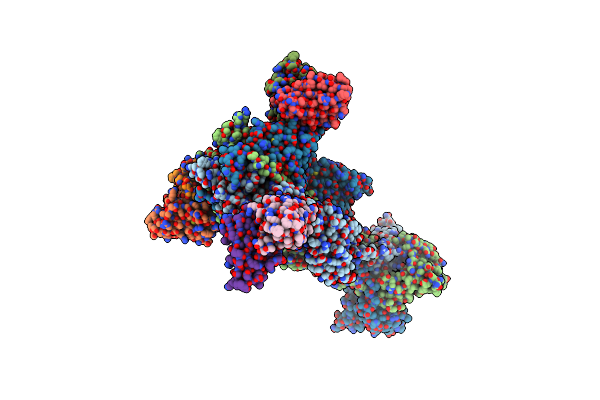
Trimeric Hsv-1F Gb Ectodomain In Postfusion Conformation With Three Bound Hdit101 Fab Molecules.
Organism: Human alphaherpesvirus 1 strain f, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.27 Å Release Date: 2024-06-19 Classification: VIRAL PROTEIN |
 |
Trimeric Hsv-1F Gb Ectodomain In Postfusion Conformation With Three Bound Hdit102 Fab Molecules.
Organism: Human alphaherpesvirus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.44 Å Release Date: 2024-06-19 Classification: VIRAL PROTEIN |
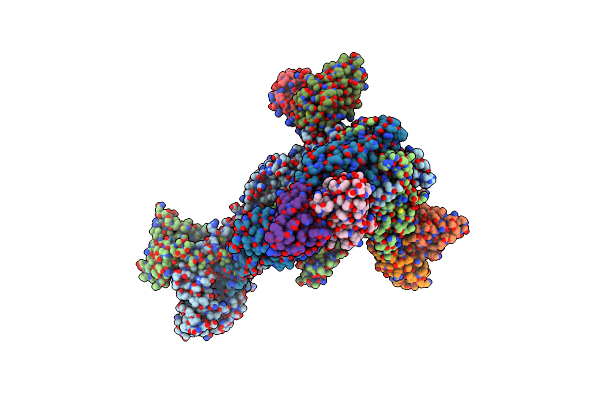 |
Trimeric Hsv-2F Gb Ectodomain In Postfusion Conformation With Three Bound Hdit101 Fab Molecules.
Organism: Human herpesvirus 2 strain g, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.45 Å Release Date: 2024-06-19 Classification: VIRAL PROTEIN |
 |
Trimeric Hsv-2G Gb Ectodomain In Postfusion Conformation With Three Bound Hdit102 Fab Molecules.
Organism: Human herpesvirus 2 strain g, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.12 Å Release Date: 2024-06-19 Classification: VIRAL PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Cryo-Em Structure Of Ca2+-Bound Mtmem16F N562A Mutant In Digitonin Closed/Closed
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: CA, P1O |
 |
Cryo-Em Structure Of Ca2+-Bound Mtmem16F N562A Mutant In Digitonin Open/Closed
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Cryo-Em Structure Of Ca2+-Bound Mtmem16F F518A Q623A Mutant In Gdn Open/Closed
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: CA |

