Search Count: 68
 |
Crystal Structure Of Susa Amylase From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, IMD, CA |
 |
Crystal Structure Of Susg From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, A1ILG, CA, IMD, ACT |
 |
Crystal Structure Of Amylase 5 (Amy5) From Ruminococcus Bromii Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Ruminococcus bromii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, A1IHI, CA |
 |
Crystal Structure Of K38 Amylase From Bacillus Sp. Strain Ksm-K38 Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacillus sp. ksm-k38
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, ACT, NA |
 |
Drosophila Golgi Alpha-Mannosidase Ii (Dgmii) In Complex With Swainsonine-Configured Alkyl Indolizidine
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2024-10-09 Classification: HYDROLASE Ligands: PEG, A1IGC, ZN |
 |
Drosophila Golgi Alpha-Mannosidase Ii (Dgmii) In Complex With Amide Modified Swainsonine-Configured Alkyl Indolizidine
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-10-09 Classification: HYDROLASE Ligands: A1IGB, EDO, ZN |
 |
Maltodextrin Phosphorylase (Malp) In Complex With A Alpha-1,2-Cyclophellitol Analogue
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-05-15 Classification: TRANSFERASE Ligands: A1H3V, EDO, PEG, SO4 |
 |
Crystal Structure Of Agd31B, Alpha-Transglucosylase, Complexed With A Non-Covalent 1,2- Cyclophellitol Aziridine
Organism: Cellvibrio japonicus ueda107
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-05-15 Classification: STRUCTURAL PROTEIN Ligands: GDQ, OXL, SO4, EDO, PG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-10 Classification: HYDROLASE Ligands: NAG, VGO, EDO, LYY, CL |
 |
Beta-Glucuronidase From Acidobacterium Capsulatum In Complex With Inhibitor R3794
Organism: Acidobacterium capsulatum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-10 Classification: HYDROLASE Ligands: VGO, PO4 |
 |
Crystal Structure Of Human Heparanase In Complex With Competitive Inhibitor Derrived From Siastatin B
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-01-10 Classification: HYDROLASE Ligands: NAG, EDO, CL, VGO |
 |
Crystal Structure Of Human Heparanase In Complex With Glucuronic Acid Configured 3-Geminal Diol Iminosugar Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-01-10 Classification: HYDROLASE Ligands: VP5, EDO, NAG, CL |
 |
Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum In Complex With Competitive Inhibitor Derrived From Siastatin B
Organism: Acidobacterium capsulatum
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2024-01-10 Classification: HYDROLASE Ligands: VON, SO4 |
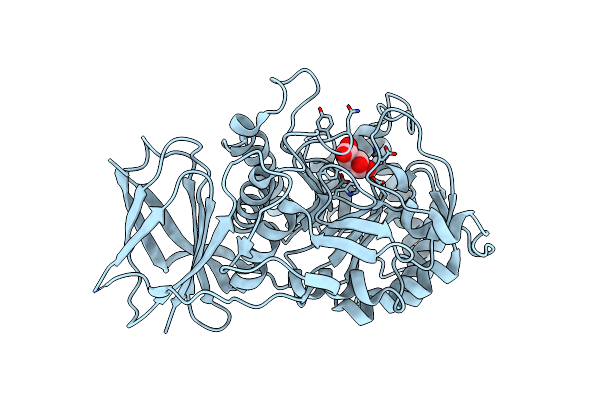 |
Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum In Complex With Glucuronic Acid Configured Isofagamine
Organism: Acidobacterium capsulatum
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-01-10 Classification: HYDROLASE Ligands: SJ5 |
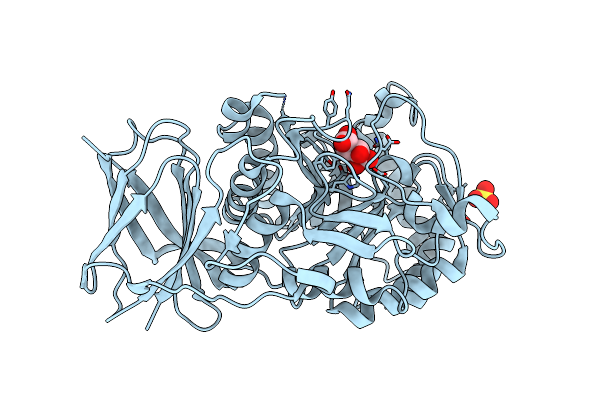 |
Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum In Complex With Glucuronic Acid Configured 3-Geminal Diol Iminosugar Inhibitor
Organism: Acidobacterium capsulatum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-01-10 Classification: HYDROLASE Ligands: VP5, SO4 |
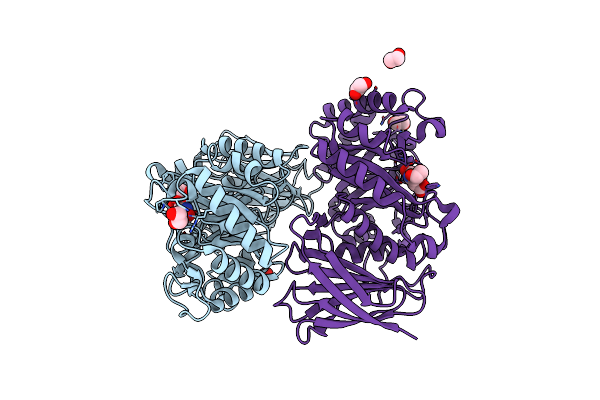 |
Crystal Structure Of Heparanase From Burkholderia Pseudomallei In Complex With Siastatin B Derived Inhibitor
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-01-10 Classification: HYDROLASE Ligands: VON, EDO |
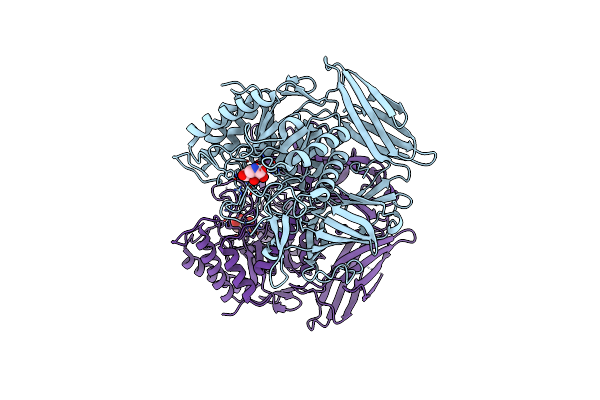 |
Crystal Structure Of Beta-Glucuronidase From Escherichia Coli In Complex With Siastatin B Derived Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: HYDROLASE Ligands: VON |
 |
Beta-L-Arabinofurano-Cyclitol Aziridines Are Cysteine-Directed Broad-Spectrum Inhibitors And Activity-Based Probes For Retaining Beta-L-Arabinofuranosidases
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-12-13 Classification: HYDROLASE Ligands: ZN, UI5 |
 |
Gh146 Beta-L-Arabinofuranosidase From Bacteroides Thetaioatomicron In Complex With Beta-L-Arabinofurano Cyclophellitol Aziridine
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-12-13 Classification: HYDROLASE Ligands: UI5, ZN, UHU |
 |
Cryo-Em Structure Of Nadh Bound Sla Dehydrogenase Rlgabd From Rhizobium Leguminosarum Bv. Trifolii Srd1565
Organism: Rhizobium leguminosarum bv. trifolii srdi565
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: NAI |

