Search Count: 9
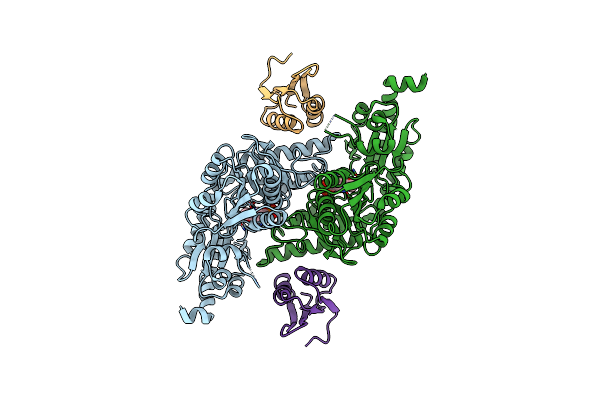 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2010-04-21 Classification: TRANSFERASE Ligands: PLP |
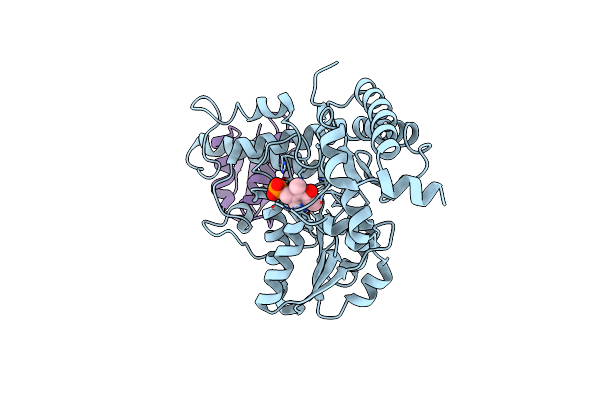 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2010-04-21 Classification: TRANSFERASE Ligands: PLP |
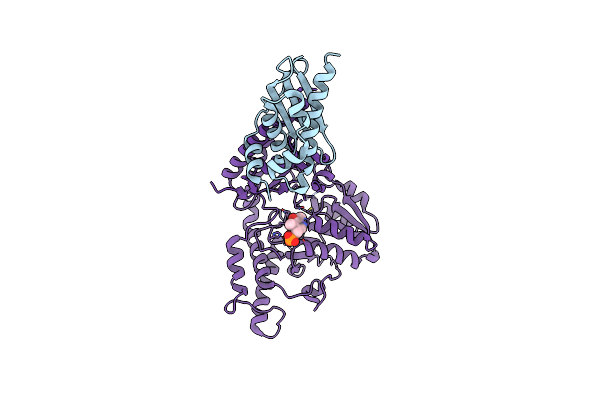 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-04-21 Classification: TRANSFERASE Ligands: PLP |
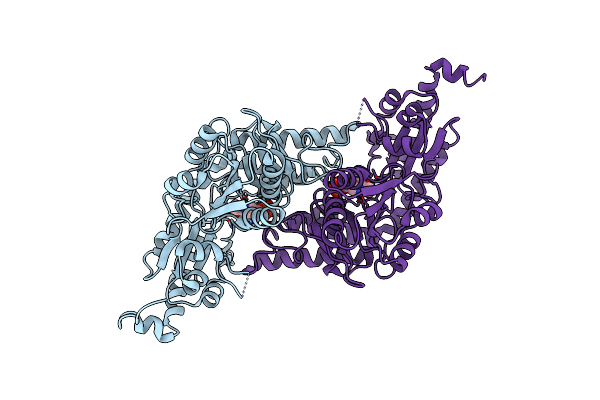 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-04-21 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Escherichia coli o157:h7 edl933
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2009-10-20 Classification: RNA BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of E.Coli Mnmg (Gida), A Highly-Conserved Trna Modifying Enzyme
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2009-03-03 Classification: RNA BINDING PROTEIN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-01-04 Classification: HYDROLASE Ligands: SO4 |
 |
Structure Of The Gtp-Binding Protein Trme From Thermotoga Maritima Complexed With 5-Formyl-Thf
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2005-01-04 Classification: HYDROLASE Ligands: FON |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2003-12-02 Classification: UNKNOWN FUNCTION |

