Search Count: 27
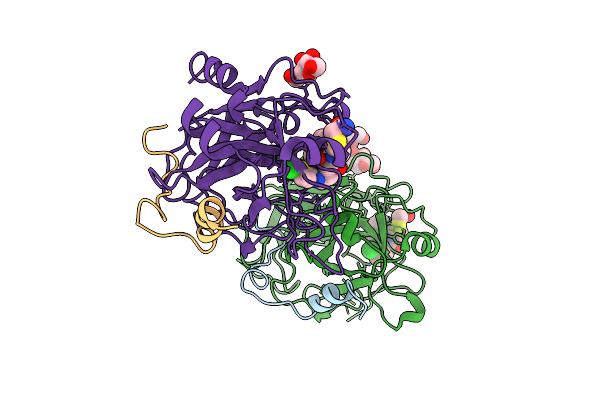 |
Crystal Structure Of Thrombin In Complex With A Chlorothiophene-Based Inhibitor, Cp3, Discovered By A Novel Rapid Nanoscale Library Screening.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1ISR, NAG |
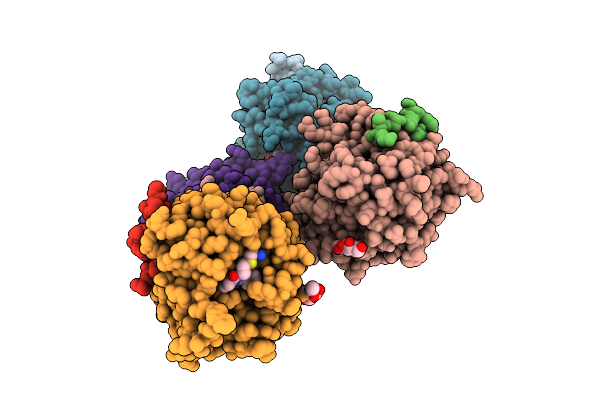 |
Crystal Structure Of Thrombin In Complex With A Chlorothiophene-Based Inhibitor, Cp2, Discovered By A Novel Rapid Nanoscale Library Screening.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1ITQ, NAG, NA |
 |
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: UNKNOWN FUNCTION Ligands: MLT |
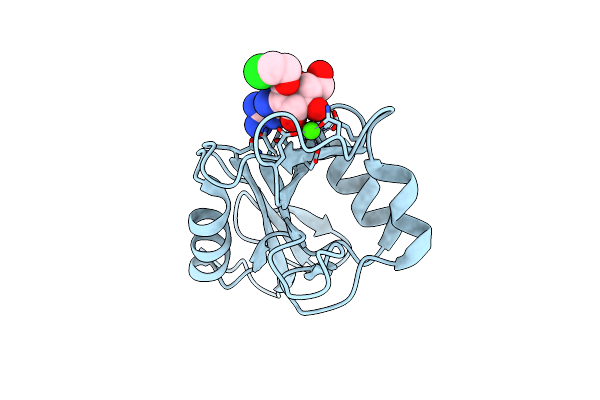 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-16 Classification: SUGAR BINDING PROTEIN Ligands: CA, CL, A1H0Q |
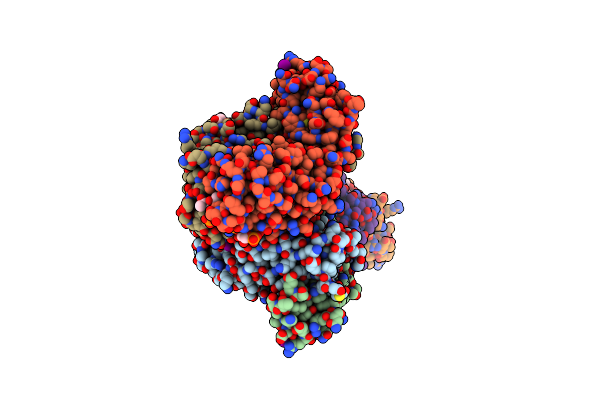 |
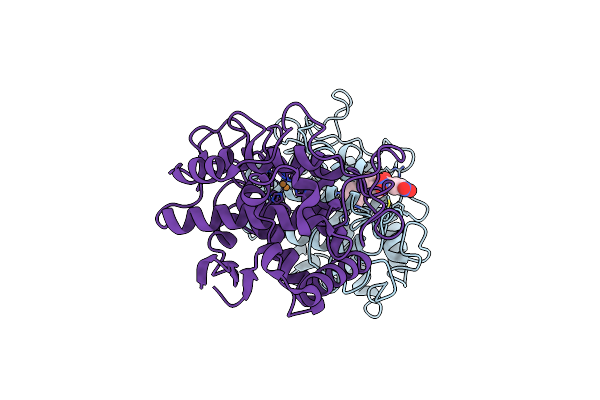
Crystal Structure Of A Neisseria Meningitidis Serogroup A Capsular Oligosaccharide Bound To A Functional Fab
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2020-10-07 Classification: IMMUNE SYSTEM Ligands: IOD, EDO, OOW, OA8, OAB |
 |
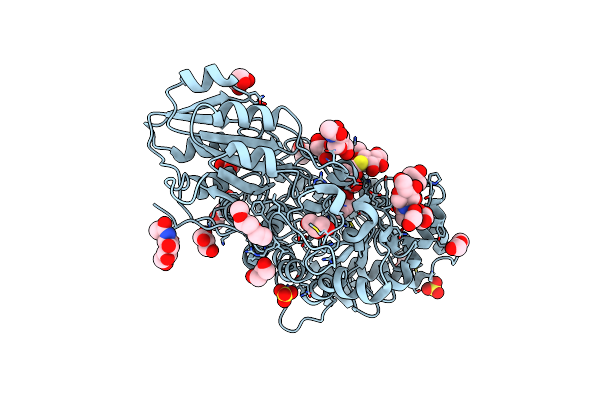
Crystal Structure Of Tyrosinase From Bacillus Megaterium With Jkb Inhibitor In The Active Site.
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-06-19 Classification: OXIDOREDUCTASE Ligands: CU, JKB |
 |
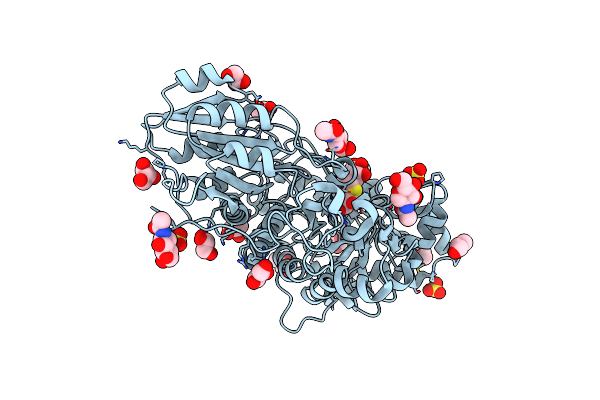
Crystal Structure Analysis Of Plant Exohydrolase In Complex With Methyl 2-Thio-Beta-Sophoroside
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2019-05-29 Classification: HYDROLASE Ligands: NAG, GOL, SO4, 1PE |
 |
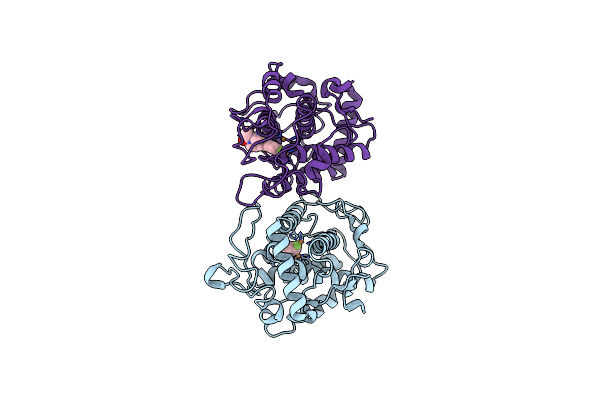
Crystal Structure Analysis Of The Variant Plant Exohydrolase Arg158Ala-Glu161Ala In Complex With Methyl 6-Thio-Beta-Gentiobioside
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-05-29 Classification: HYDROLASE Ligands: NAG, GOL, SO4 |
 |
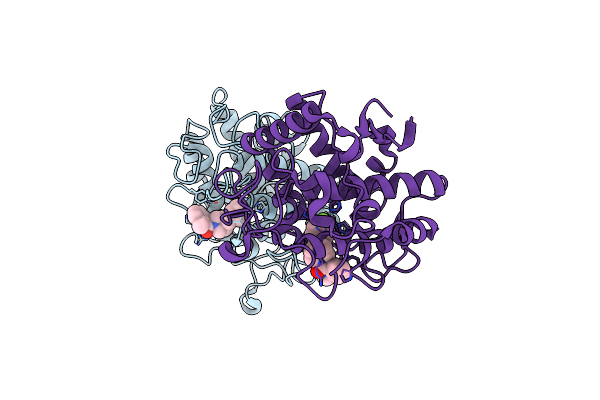
Crystal Structure Of Tyrosinase From Bacillus Megaterium With Svf Inhibitor In The Active Site
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: CU, SVF |
 |
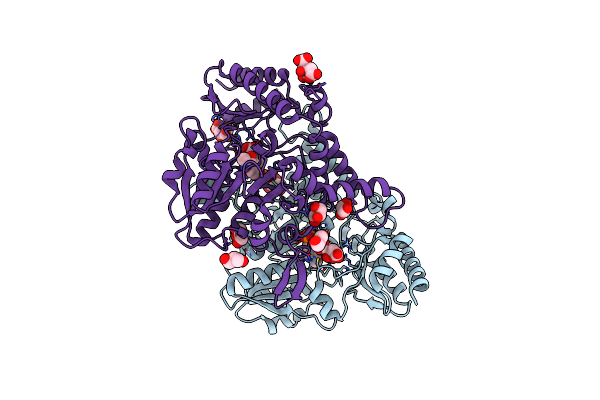
Crystal Structure Of Tyrosinase From Bacillus Megaterium With B5N Inhibitor In The Active Site
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: B5N, CU |
 |
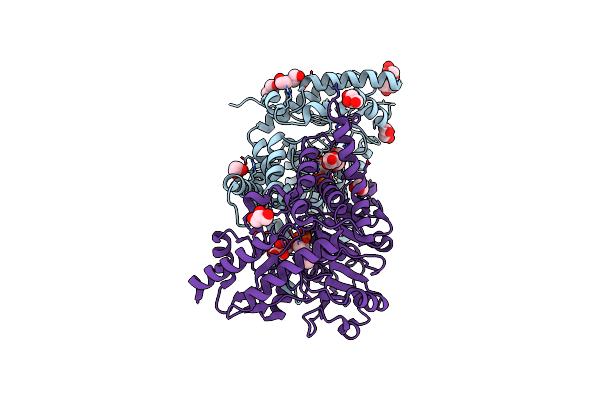
Crystal Structure Of The Cysteine Desulfurase Csda From Escherichia Coli At 1.996 Angstroem Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-12-21 Classification: TRANSFERASE Ligands: PLP, GOL, CIT |
 |
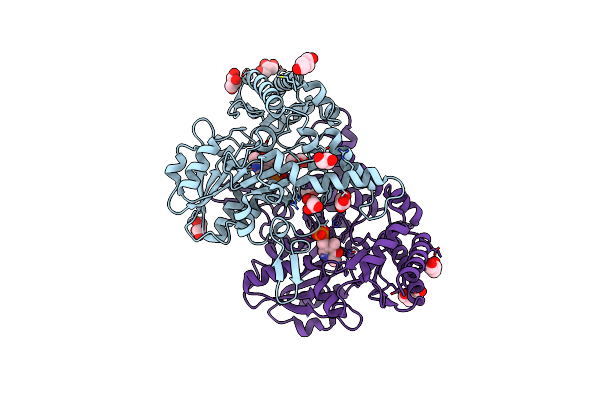
Crystal Structure Of The Cysteine Desulfurase Csda (Persulfurated) From Escherichia Coli At 2.384 Angstroem Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2016-11-23 Classification: TRANSFERASE Ligands: GOL, PEG, TLA, PLP |
 |
Crystal Structure Of The Cysteine Desulfurase Csda (S-Sulfonic Acid) From Escherichia Coli At 2.050 Angstroem Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-11-23 Classification: TRANSFERASE Ligands: GOL, PEG, PLP |
 |
Crystal Structure Of The Complex Between The Cysteine Desulfurase Csda And The Sulfur-Acceptor Csde In The Persulfurated State At 2.50 Angstroem Resolution
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-11-23 Classification: TRANSFERASE Ligands: PLP, PEG, GOL |
 |
Crystal Structure Of E. Coli Trna N6-Threonylcarbamoyladenosine Dehydratase, Tcda, In Complex With Atp At 1.768 Angstroem Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-05-06 Classification: LIGASE Ligands: K, ATP, GOL, NA |
 |
Crystal Structure Of E. Coli Trna N6-Threonylcarbamoyladenosine Dehydratase, Tcda, In Complex With Amp At 1.801 Angstroem Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-05-06 Classification: LIGASE Ligands: K, AMP, GOL, NA, PO4 |
 |
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-03-25 Classification: HYDROLASE Ligands: BGC, GOL, SO4 |
 |
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-03-25 Classification: HYDROLASE Ligands: NAG, GOL, SO4 |
 |
Crystal Structure Of Barley Beta-D-Glucan Glucohydrolase Isoenzyme Exo1 In Complex With 3-Deoxy-Glucose
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2015-03-25 Classification: HYDROLASE Ligands: BGC, 3DO, GOL, SO4 |
 |
Crystal Structure Of Barley Beta-D-Glucan Glucohydrolase Isoenzyme Exo1 In Complex With 4-Deoxy-Glucose
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-25 Classification: HYDROLASE Ligands: BGC, GOL, SO4 |

