Search Count: 4
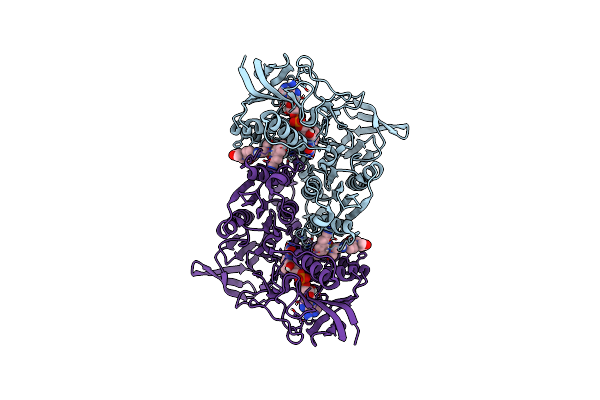 |
Cryo-Em Structure Of Mtb Lpd Bound To Inhibitor Complex With 2-((2-Cyano-N,5-Dimethyl-1H-Indole)-7-Sulfonamido)-N-(4-(Oxetan-3-Yl)-3,4-Dihydro-2H-Benzo[B] [1,4]Oxazin-7-Yl)Acetamide
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: FAD, OU6 |
 |
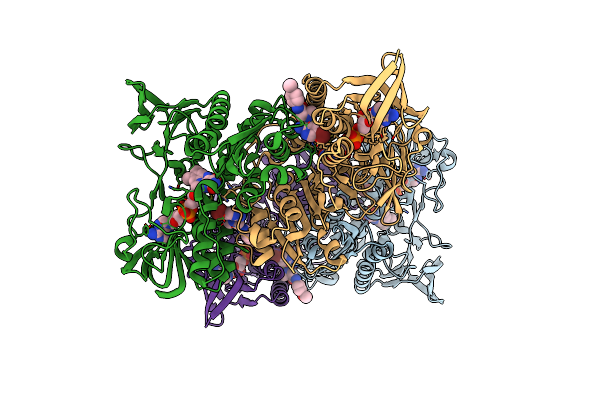
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2021-01-27 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, WPM, GOL |
 |
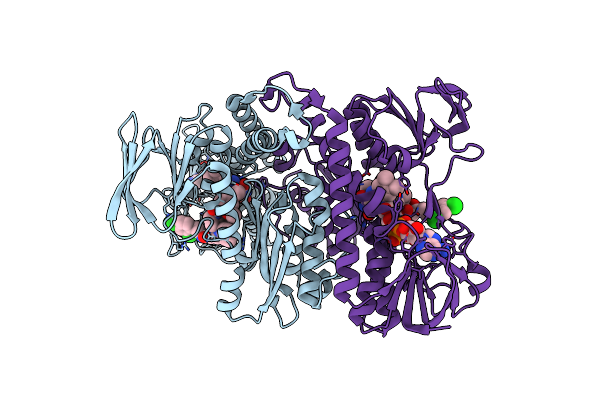
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, M52 |
 |
Structure Of Mycobacterial Lipoamide Dehydrogenase Bound To A Triazaspirodimethoxybenzoyl Inhibitor
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2010-01-26 Classification: OXIDOREDUCTASE Ligands: FAD, 3II |

