Search Count: 35
 |
Crystal Structure Of Chitinase (E167Q) From The Carnivorous Plant Drosera Adelae
Organism: Drosera adelae
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: ACY |
 |
Organism: Drosera adelae
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: NAG, ACY |
 |
Organism: Gallus gallus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-05-18 Classification: DE NOVO PROTEIN Ligands: MG |
 |
Crystal Structure Of Fmn-Dependent Nadph-Quinone Reductase (Azor) From Bacillus Cohnii
Organism: Bacillus cohnii
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: FMN, IPA, GOL |
 |
Organism: Parvibaculum lavamentivorans ds-1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-02-02 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Indigo Reductase From Bacillus Smithii Type Strain Dsm 4216
Organism: Bacillus smithii
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2020-04-01 Classification: OXIDOREDUCTASE Ligands: FMN, PE8, NHE |
 |
Crystal Structure Of Indigo Reductase (Y151F) From Bacillus Smithii Type Strain Dsm 4216
Organism: Bacillus smithii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-04-01 Classification: OXIDOREDUCTASE Ligands: FMN, PE8 |
 |
Organism: Phytophthora infestans (strain t30-4)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2020-04-01 Classification: OXIDOREDUCTASE Ligands: FLC, PE8, NAD |
 |
Structure Of A Glycoside Hydrolase Family 3 Beta-N-Acetylglucosaminidase From Paenibacillus Sp. Str. Fpu-7
Organism: Paenibacillaceae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-09-25 Classification: HYDROLASE Ligands: NAG, GOL |
 |
Crystal Structure Of The Nadp+ And Tartrate-Bound Complex Of L-Serine 3-Dehydrogenase From The Hyperthermophilic Archaeon Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2018-02-07 Classification: OXIDOREDUCTASE Ligands: TLA, NAP, ACY, SO4 |
 |
Organism: Aeropyrum pernix (strain atcc 700893 / dsm 11879 / jcm 9820 / nbrc 100138 / k1)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2016-06-15 Classification: OXIDOREDUCTASE Ligands: NDP |
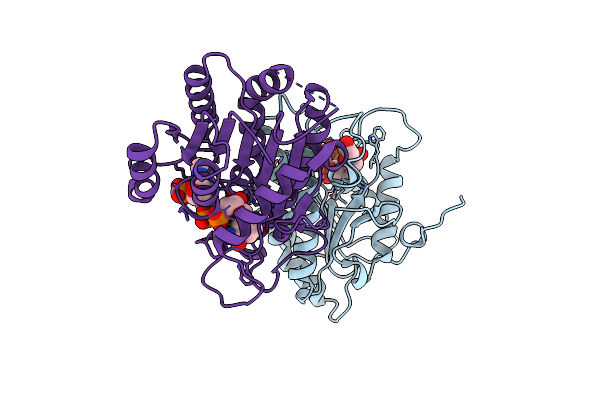 |
Crystal Structure Of Nadph Bound Carbonyl Reductase From Chicken Fatty Liver
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2015-07-29 Classification: OXIDOREDUCTASE ACTIVATOR Ligands: NDP, EDO |
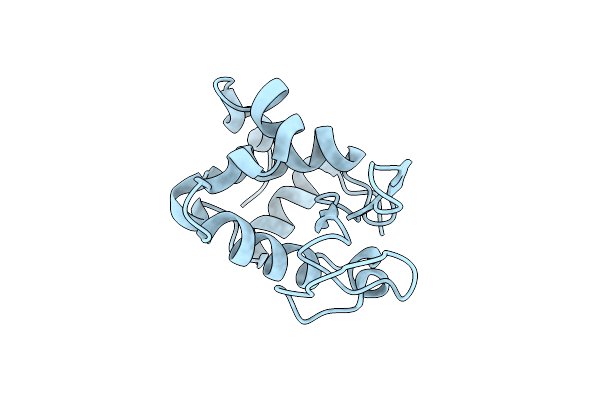 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2015-06-10 Classification: HYDROLASE |
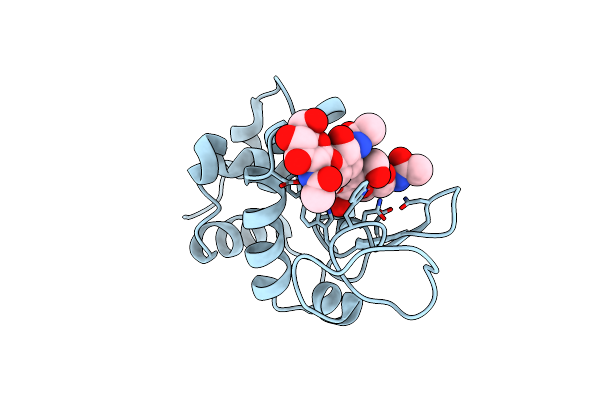 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2015-06-10 Classification: HYDROLASE |
 |
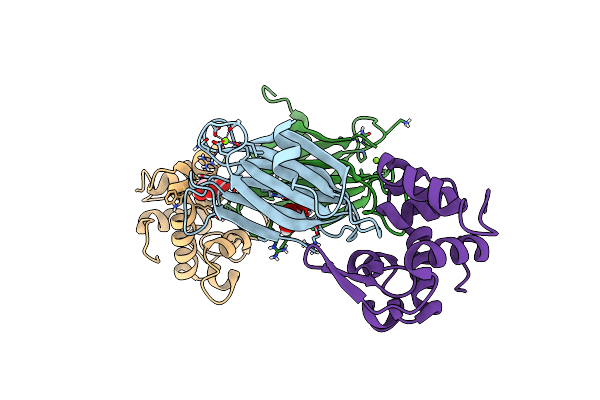
The 1.18 A Resolution Structure Of L-Serine 3-Dehydrogenase Complexed With Nadp+ And Sulfate Ion From The Hyperthermophilic Archaeon Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2015-03-04 Classification: OXIDOREDUCTASE Ligands: NAP, SO4, ACY |
 |
Crystal Structure Of Aeromonas Hydrophila Plii In Complex With Meretrix Lusoria Lysozyme
Organism: Aeromonas hydrophila subsp. hydrophila, Meretrix lusoria
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2015-02-11 Classification: Hydrolase/Hydrolase inhibitor Ligands: GOL, MG |
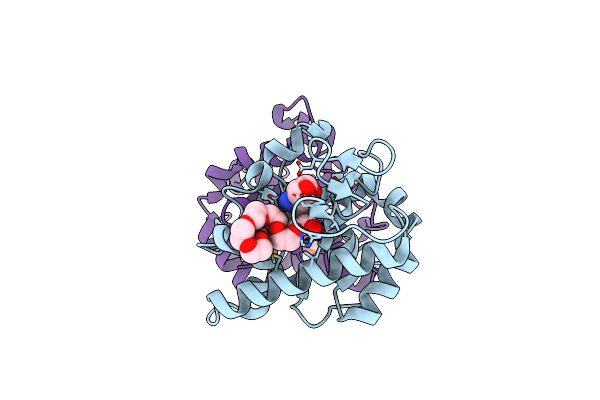 |
Organism: Struthio camelus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-10-15 Classification: HYDROLASE Ligands: TAM, P6G |
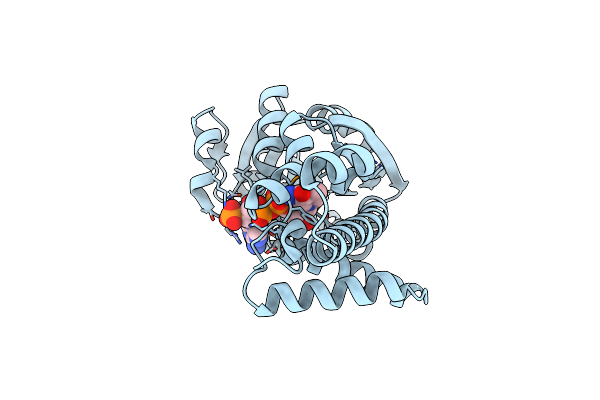 |
Crystal Structure Of Nadp Bound L-Serine 3-Dehydrogenase From Hyperthermophilic Archaeon Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: NAP |
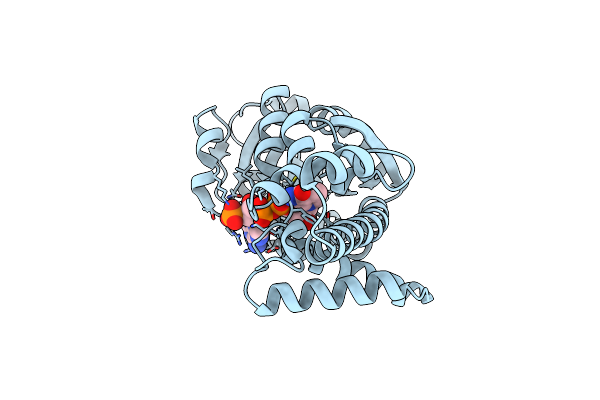 |
Crystal Structure Of Nadp Bound L-Serine 3-Dehydrogenase (K170M) From Hyperthermophilic Archaeon Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: NAP |

