Search Count: 14
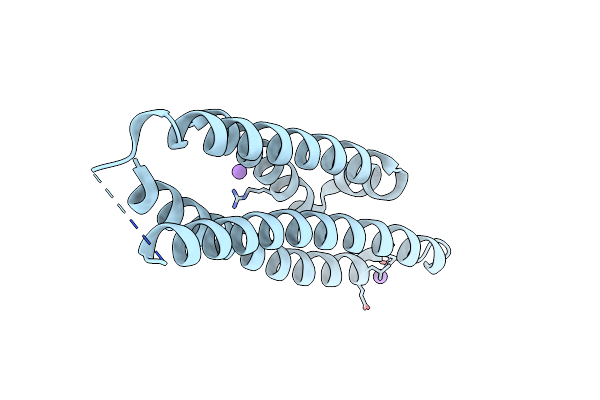 |
Crystal Structure Of The N-Terminal Domain Of Mutants Of Human Apolipoprotein-E (Apoe)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-07-20 Classification: LIPID TRANSPORT |
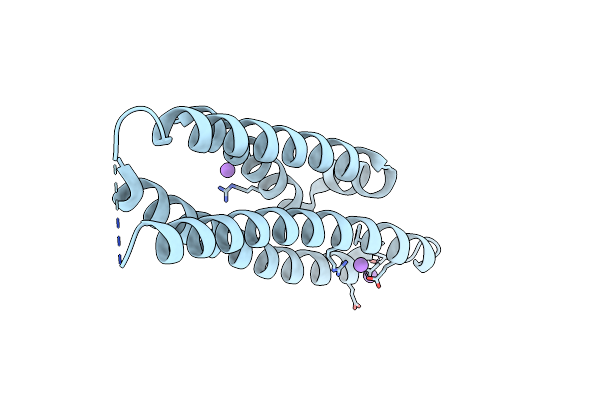 |
Crystal Structure Of The N-Terminal Domain Of Mutants Of Human Apolipoprotein-E (Apoe)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-20 Classification: LIPID TRANSPORT |
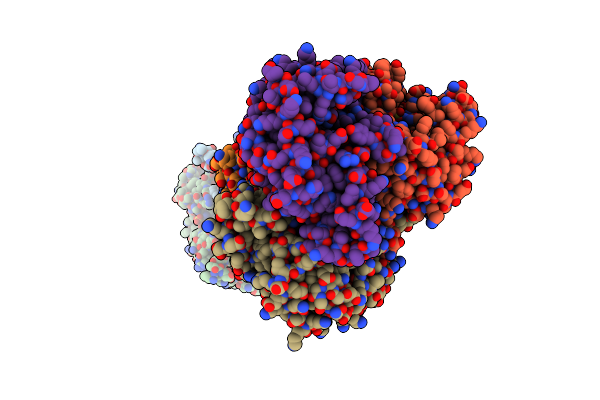 |
Re-Refined Structure Of R-State L-Lactate Dehydrogenase Fromlactobacillus Casei
Organism: Lactobacillus casei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE Ligands: SO4 |
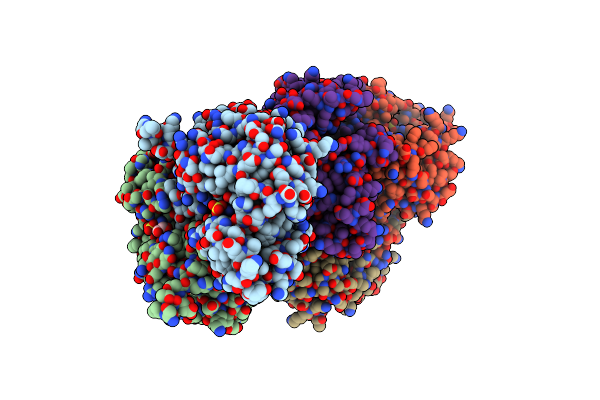 |
Organism: Lactobacillus casei subsp. casei atcc 393
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-02-06 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
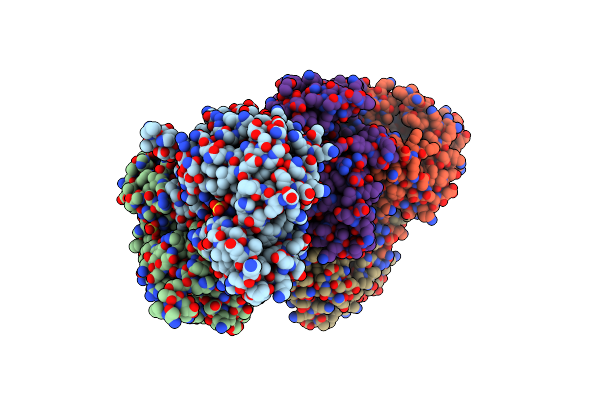
Complex Structure Of Lactobacillus Casei Lactate Dehydrogenase With Fructose-1,6-Bisphosphate
Organism: Lactobacillus casei subsp. casei atcc 393
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-02-06 Classification: OXIDOREDUCTASE Ligands: SO4, FBP |
 |
Complex Structure Of Lactobacillus Casei Lactate Dehydrogenase Penta Mutant With Pyruvate
Organism: Lactobacillus casei subsp. casei atcc 393
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2019-02-06 Classification: OXIDOREDUCTASE Ligands: PYR, SO4 |
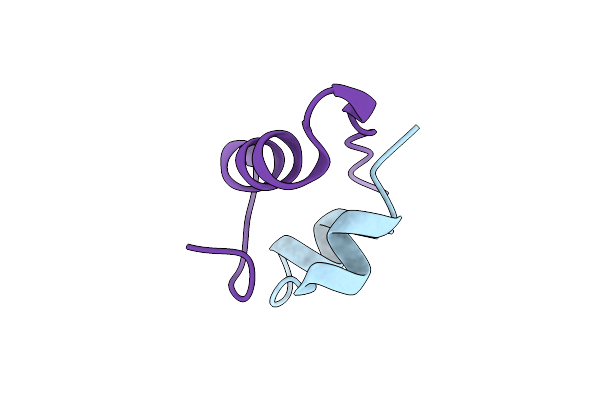 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-05-03 Classification: HORMONE |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-07-23 Classification: OXIDOREDUCTASE |
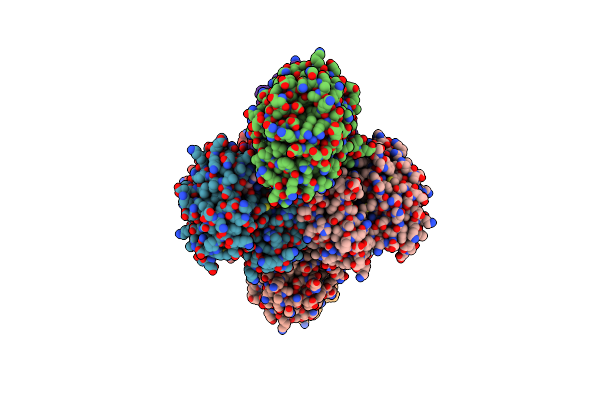 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-07-23 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Thermus caldophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-03-06 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
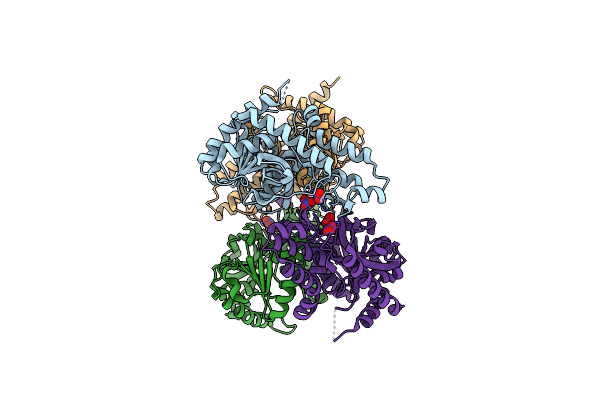
L-Lactate Dehydrogenase From Thermus Caldophilus Gk24 Complexed With Oxamate, Nadh And Fbp
Organism: Thermus caldophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-03-06 Classification: OXIDOREDUCTASE Ligands: OXM, FBP, NAD, GOL |
 |
T-State Structure Of Allosteric L-Lactate Dehydrogenase From Lactobacillus Casei
Organism: Lactobacillus casei
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2009-09-08 Classification: OXIDOREDUCTASE Ligands: NO3 |
 |
R-State Structure Of Allosteric L-Lactate Dehydrogenase From Lactobacillus Casei
Organism: Lactobacillus casei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-09-08 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Organism: Gymnema sylvestre
Method: SOLUTION NMR Release Date: 1996-08-01 Classification: SWEET TASTE-SUPPRESSING PROTEIN |

