Search Count: 31
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL, PEG |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: A1L6P, NA |
 |
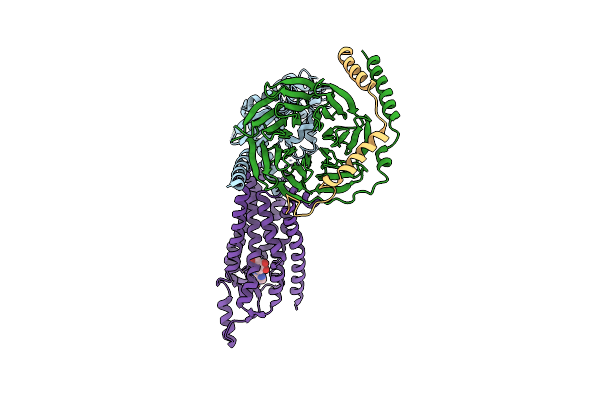
Cryo-Em Structure Of The Human Adenosine A1 Receptor-Gi2-Protein Complex Bound To Its Endogenous Agonist And An Allosteric Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: ADN, XTD |
 |
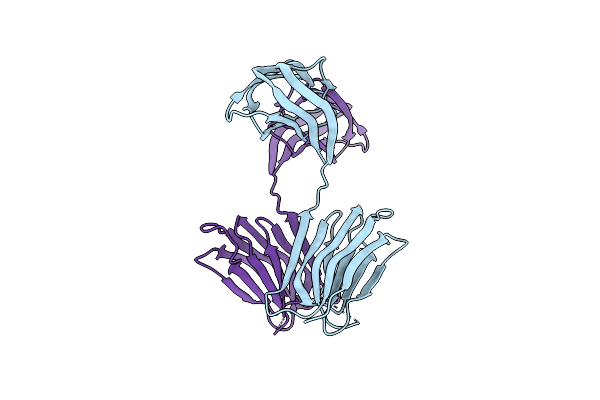
Cryo-Em Structure Of The Human Adenosine A1 Receptor-Gi2-Protein Complex Bound To Its Endogenous Agonist
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: ADN |
 |
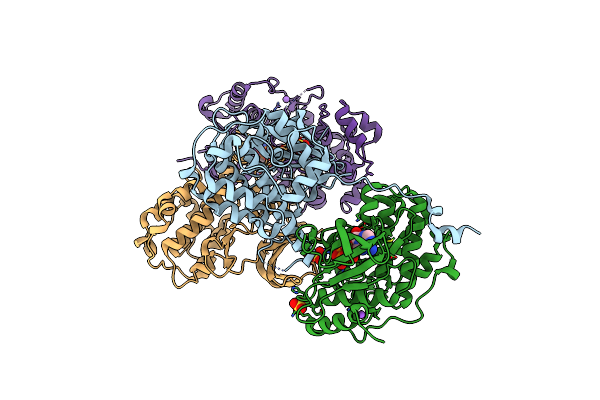
Structure Determination Of A223, A Turret Protein In Sulfolobus Turreted Icosahedral Virus, Using An Iterative Hybrid Approach
Organism: Sulfolobus turreted icosahedral virus 1
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2018-11-21 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-11-12 Classification: TRANSFERASE Ligands: ANP, SO4, NA |
 |
Life In The Extremes: Atomic Structure Of Sulfolobus Turreted Icosahedral Virus
Organism: Sulfolobus turreted icosahedral virus
Method: ELECTRON MICROSCOPY Release Date: 2013-05-01 Classification: VIRUS |
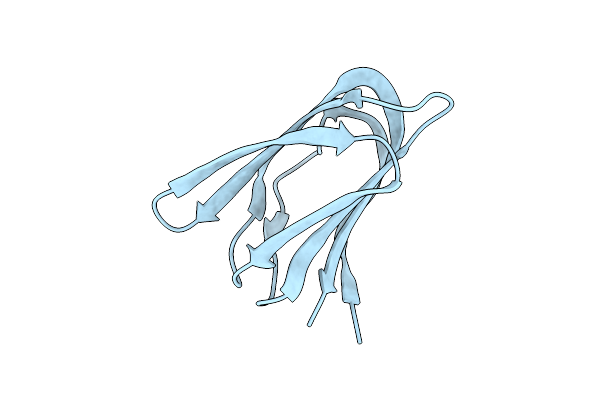 |
Crystal Structure Of A223 C-Terminal Domain, A Structural Protein From Sulfolobus Turreted Icosahedral Virus (Stiv)
Organism: Sulfolobus turreted icosahedral virus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-04-03 Classification: VIRAL PROTEIN |
 |
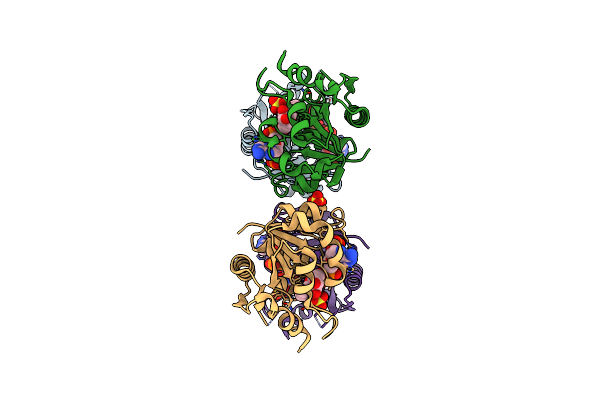
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-10-31 Classification: TRANSFERASE/SIGNALING PROTEIN Ligands: ANP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-08-15 Classification: TRANSFERASE Ligands: ANP |
 |
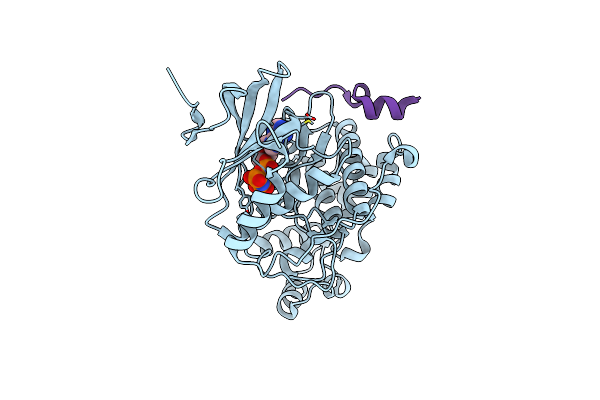
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-05-30 Classification: OXIDOREDUCTASE Ligands: NAP, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-02-29 Classification: TRANSFERASE Ligands: ANP |
 |
Organism: Homo sapiens, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-02-22 Classification: TRANSFERASE |
 |
Linear Binding Motifs For Jnk And For Calcineurin Antagonistically Control The Nuclear Shuttling Of Nfat4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2011-09-28 Classification: TRANSCRIPTION Ligands: ANP, GOL |
 |
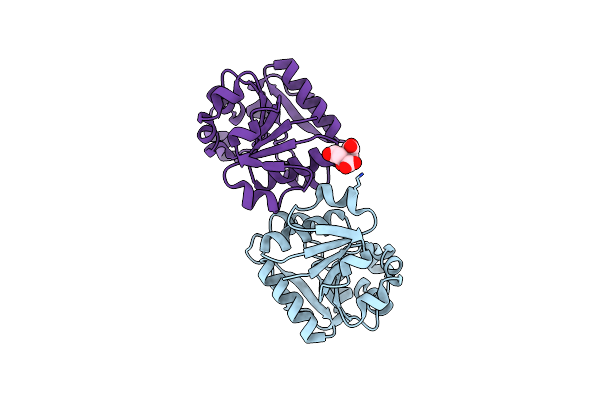
Linear Binding Motifs For Jnk And For Calcineurin Antagonistically Control The Nuclear Shuttling Of Nfat4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-09-28 Classification: TRANSCRIPTION Ligands: ANP |
 |
Crystal Structure Of The Membrane Proximal Oxidoreductase Domain Of Human Steap3, The Dominant Ferric Reductase Of The Erythroid Transferrin Cycle
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-05-06 Classification: OXIDOREDUCTASE Ligands: CIT |
 |
Crystal Structure Of The Membrane Proximal Oxidoreductase Domain Of Human Steap3, The Dominant Ferric Reductase Of The Erythroid Transferrin Cycle
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-05-06 Classification: OXIDOREDUCTASE Ligands: NAP, CIT |
 |
Three-Dimensional Crystal Structure Of Dissimilatory Sulfite Reductase D (Dsrd)
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2003-10-14 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2003-07-03 Classification: TRANSCRIPTION |

