Search Count: 27
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE Ligands: CYS, ZN, CU |
 |
Crystal Structure Of Ketosteroid Isomerase Variant M105A From Pseudomonos Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2011-08-03 Classification: ISOMERASE |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-03-23 Classification: LYASE Ligands: 3CO, GOL |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Beta-E56Q From Pseudomonas Putida.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-03-23 Classification: LYASE Ligands: GOL, 3CO |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Beta-H71L From Pseudomonas Putida.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-03-23 Classification: LYASE Ligands: 3CO |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Alpha-E168Q From Pseudomonas Putida.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-03-23 Classification: LYASE Ligands: GOL, 3CO |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Beta-Y215F From Pseudomonas Putida.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-03-23 Classification: LYASE Ligands: GOL, 3CO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-11-24 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: 842 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-09-22 Classification: HYDROLASE Ligands: 957 |
 |
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-02-02 Classification: OXIDOREDUCTASE Ligands: IMP, C64, ACT |
 |
Design And Synthesis Of Potent Bace-1 Inhibitors With Cellular Activity: Structure-Activity Relationship Of P1 Substituents
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-01-05 Classification: HYDROLASE Ligands: 1LI |
 |
Design And Synthesis Of Potent Bace-1 Inhibitors With Cellular Activity: Structure-Activity Relationship Of P1 Substituents
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-01-05 Classification: HYDROLASE Ligands: SO4, GOL, 2LI |
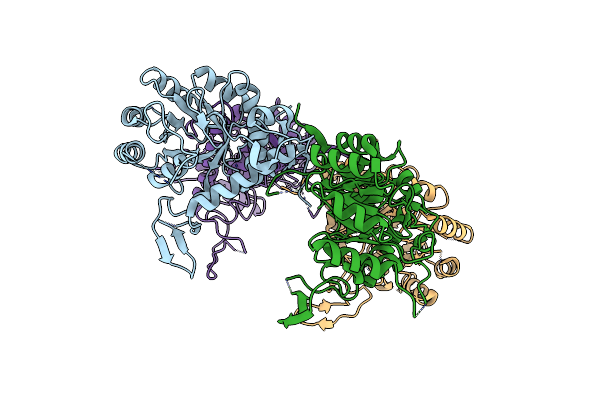 |
The Crystal Structure Of Cryptosporidium Parvum Inosine-5'-Monophosphate Dehydrogenase
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2009-12-15 Classification: OXIDOREDUCTASE |
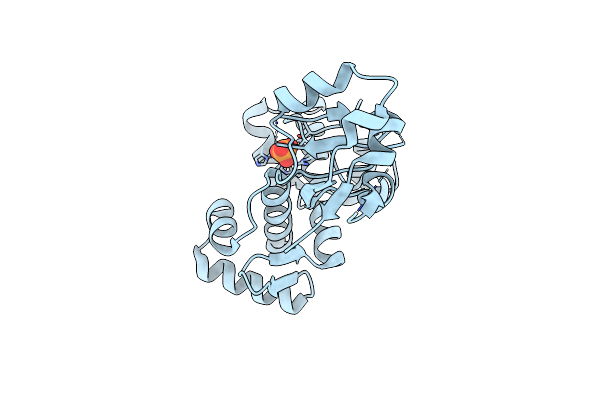 |
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-06-09 Classification: TRANSCRIPTION Ligands: NI, PO4 |
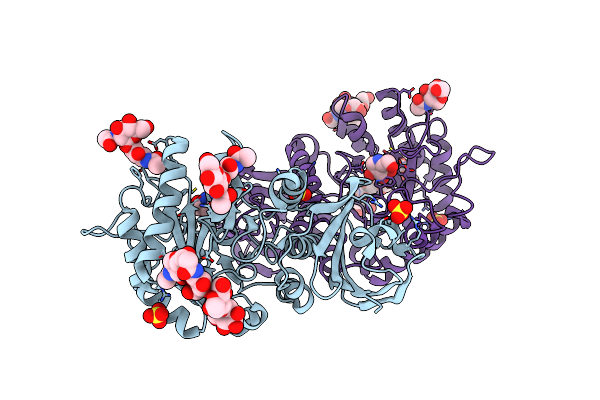 |
Crystal Structure Of Alpha-Galactosidase A At Ph 4.5 Complexed With 1-Deoxygalactonijirimycin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-05-26 Classification: HYDROLASE Ligands: SO4, NOJ, NAG |
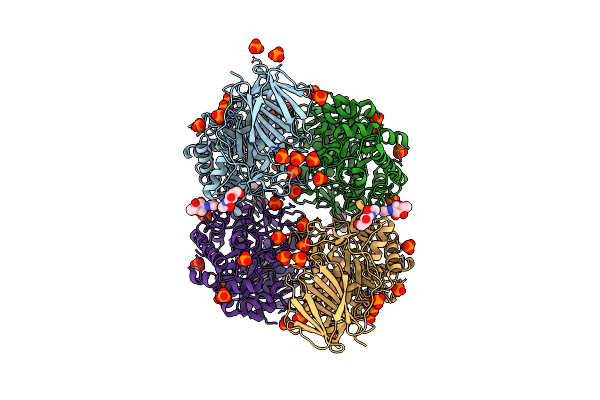 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-05-05 Classification: HYDROLASE Ligands: PO4, NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-05-05 Classification: HYDROLASE Ligands: PO4, NAG, GOL, IFM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2009-05-05 Classification: HYDROLASE Ligands: NAG, PO4 |
 |
Crystal Structure Of Acid-Beta-Glucosidase At Ph 4.5, Phosphate Crystallization Condition
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-05-05 Classification: HYDROLASE Ligands: NAG, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2009-05-05 Classification: HYDROLASE Ligands: SO4 |

