Search Count: 1,449
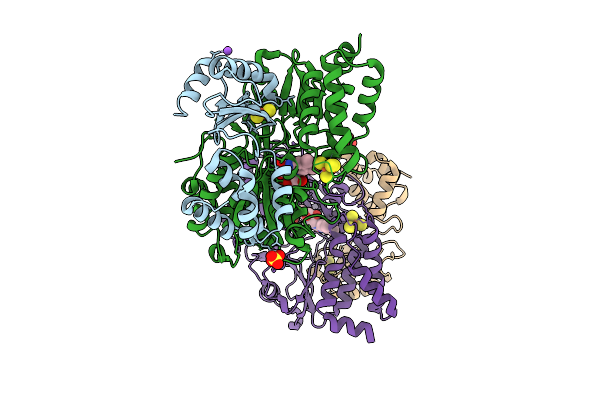 |
Crystal Structure Of The Oxidized Respiratory Complex I Subunit Nuoef From Aquifex Aeolicus, Mutation V90P(Nuoe)
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FES, NA, SO4, FMN, SF4 |
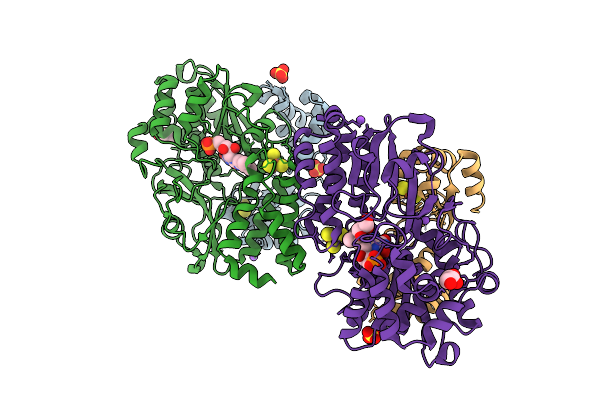 |
Crystal Structure Of The Oxidized Respiratory Complex I Subunit Nuoef From Aquifex Aeolicus, Mutation V136M(Nuoe)
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FES, CL, SO4, SF4, FMN, GOL, NA |
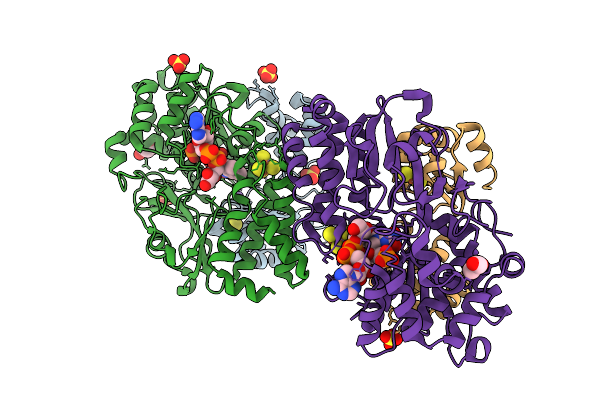 |
Crystal Structure Of The Oxidized Respiratory Complex I Subunit Nuoef From Aquifex Aeolicus, Mutation V136M(Nuoe), Bound To Nad+
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FES, CL, SO4, NA, SF4, FMN, NAD, GOL |
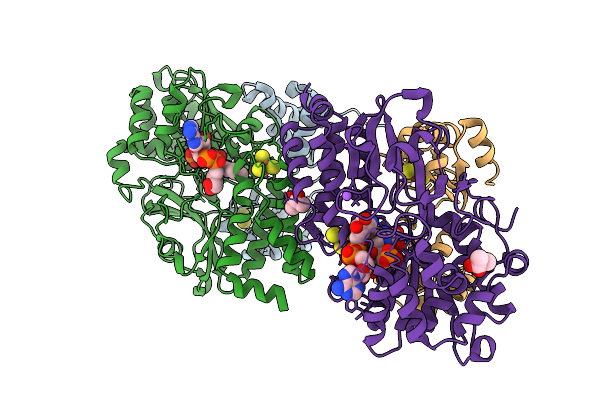 |
Crystal Structure Of The Oxidized Respiratory Complex I Subunit Nuoef From Aquifex Aeolicus, Double Mutation V90P And V136M(Nuoe), Bound To Nad+
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FES, SF4, FMN, NAD, NA, BU3, CL |
 |
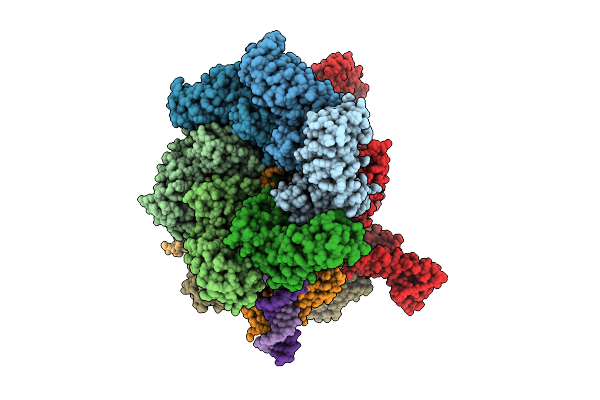
De Novo Sign Rna Polymerase Transcription Initiation Intermediate With Pre-Catalytic Bebp State (Rpi1 Open Ring)
Organism: Aquifex aeolicus vf5, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSCRIPTION Ligands: ATP, MG, POP, ZN |
 |
De Novo Sign Rna Polymerase Transcription Initiation Intermediate With Post-Catalytic Bebp State (Rpi1 Closed Ring)
Organism: Aquifex aeolicus vf5, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSCRIPTION Ligands: ATP, MG, ADP, POP, ZN |
 |
De Novo Sign Rna Polymerase Transcription Initiation Intermediate With Bound Sign-Rii
Organism: Aquifex aeolicus vf5, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSCRIPTION, TRANSFERASE/DNA Ligands: ATP, MG, ADP, POP, ZN |
 |
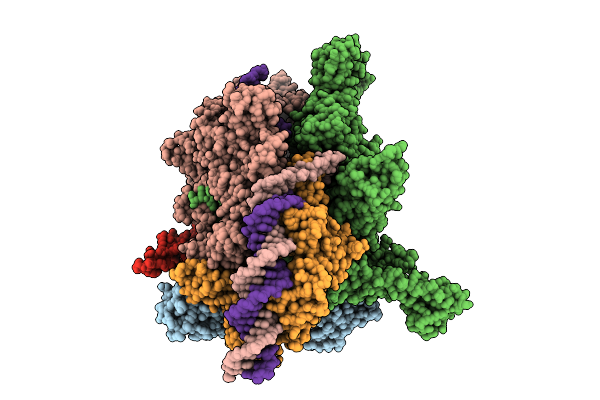
Organism: Escherichia coli, Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSCRIPTION Ligands: POP, MG, ZN |
 |
Organism: Escherichia coli, Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSCRIPTION Ligands: MG, ZN, ATP, ADP |
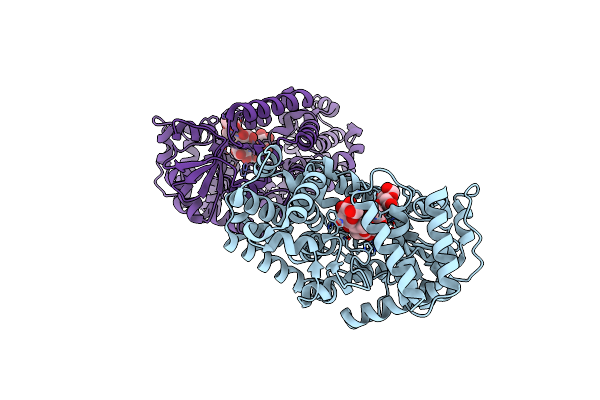 |
Crystal Structure Of Gh57 Family Amylopullulanase Mutant D352N From Aquifex Aeolicus In Complex Wih 6-Alpha-D-Maltotriosyl-Maltotriose
Organism: Aquifex aeolicus (strain vf5)
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2025-06-04 Classification: HYDROLASE |
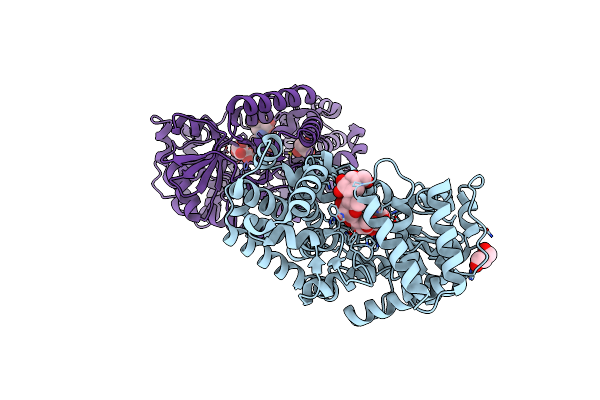 |
Crystal Structure Of Gh57 Family Amylopullulanase From Aquifex Aeolicus In Complex With Acarbose
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Aquifex aeolicus (strain vf5)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-06-04 Classification: HYDROLASE |
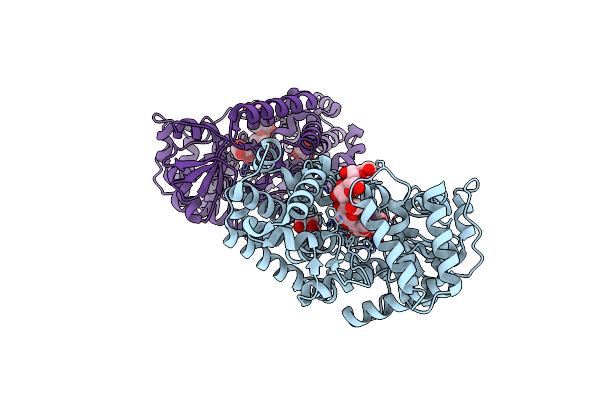 |
Crystal Structure Of Gh57 Family Amylopullulanase From Aquifex Aeolicus Mutant D352N In Complex With Maltopentaose
Organism: Aquifex aeolicus (strain vf5)
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: GOL |
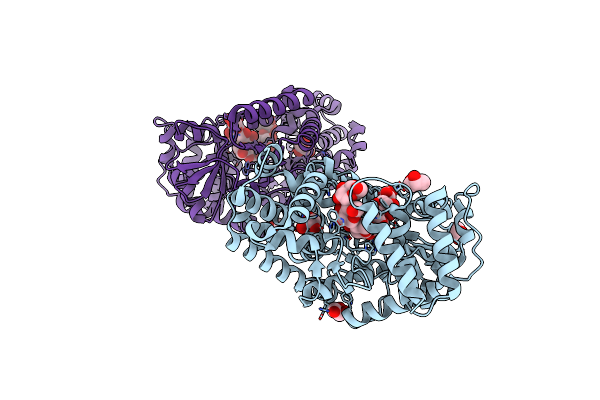 |
Crystal Structure Of Gh57 Family Amylopullulanase Mutant D352N From Aquifex Aeolicusin Complex With Maltohexaose
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: GOL |
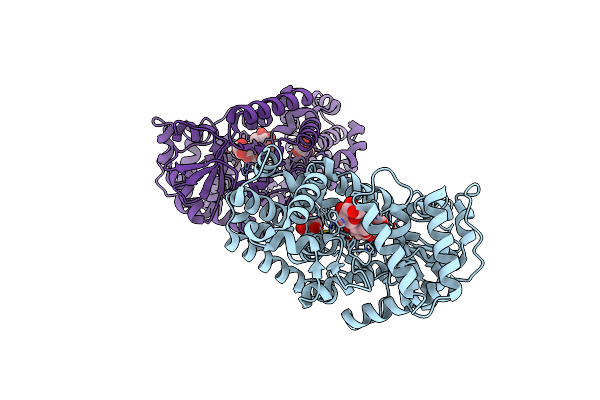 |
Co-Crystal Structure Of Gh57 Family Amylopullulanase Wild Type From Aquifex Aeolicus With Maltopentaose
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: GOL |
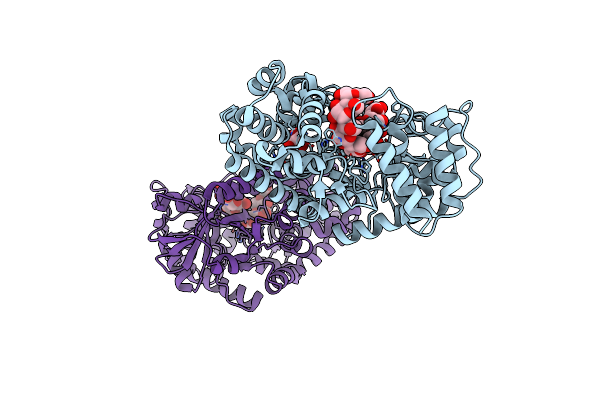 |
Crystal Structure Of Gh57 Family Amylopullulanase Mutant D352N From Aquifex Aeolicus In Complex With Maltooctaose
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: GOL |
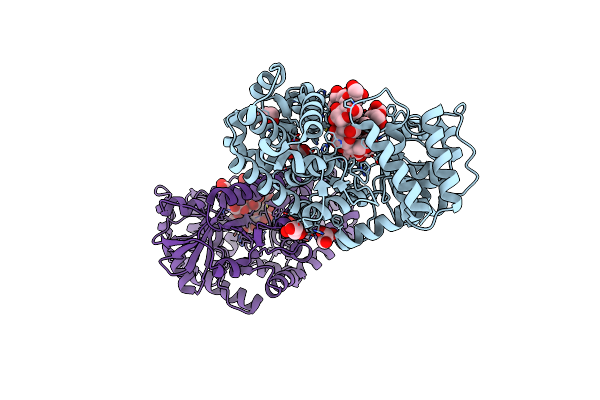 |
Gh57 Family Amylopullulanase Mutant E256L From Aquifex Aeolicus In Complex With Maltohexaose
Organism: Aquifex aeolicus (strain vf5)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: GOL |
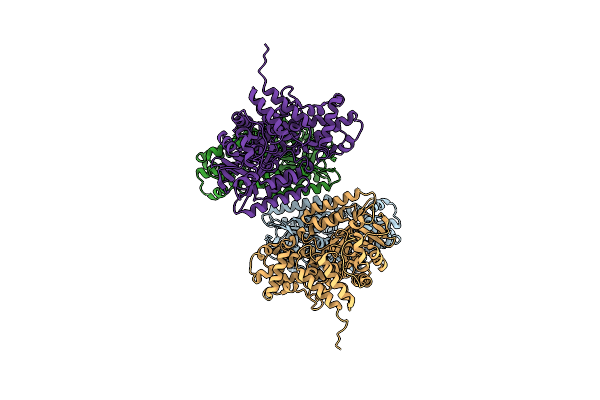 |
Crystal Structure Of Gh57 Family Amylopullulanase From Aquifex Aeolicus Wild Type Apo
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-06-04 Classification: HYDROLASE |
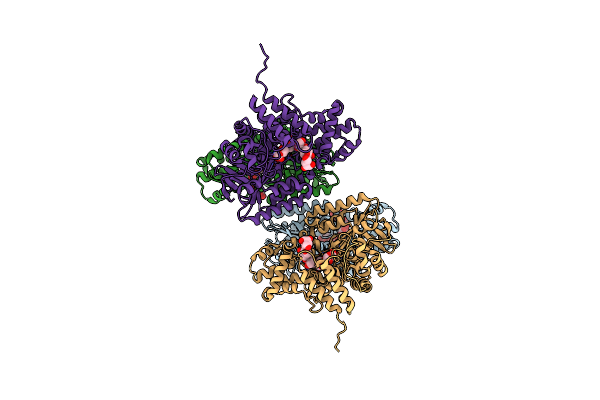 |
Crystal Structure Of Gh57 Family Amylopullulanase From Aquifex Aeolicus Mutant E256Q In Complex With Maltopentaose
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2025-06-04 Classification: HYDROLASE |
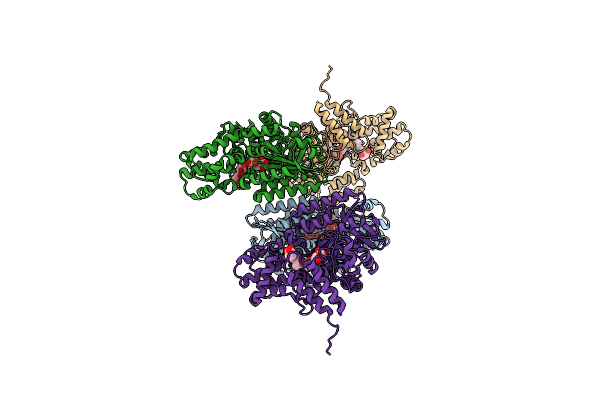 |
Crystal Structure Of Gh57 Family Amylopullulanase From Aquifex Aeolicus Wild Type In Complex With Acarbose
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2025-06-04 Classification: HYDROLASE |

