Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Remdesivir Triphosphate, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: NWX, MG, ZN |
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Atp, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: ATP, MG, ZN |
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Utp, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: ZN, MG, UTP |
 |
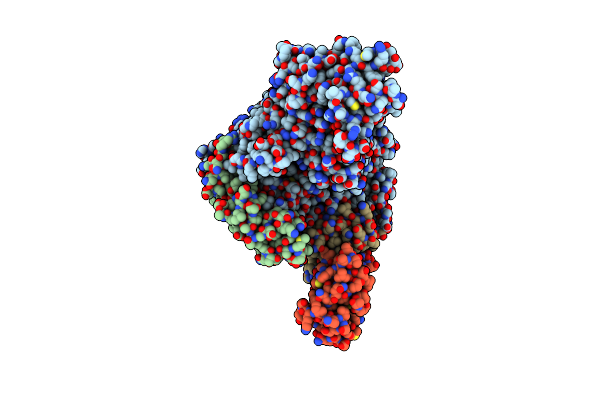
Sars-Cov-2 Replication-Transcription Complex Bound To Gtp, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: ZN, MG, GTP, L2B |
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Ctp, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: ZN, MG, CTP, L2B |
 |
Crystal Structure Of The Hiv Capsid Hexamer Bound To The Small Molecule Long-Acting Inhibitor, Gs-6207
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN Ligands: QNG |
 |
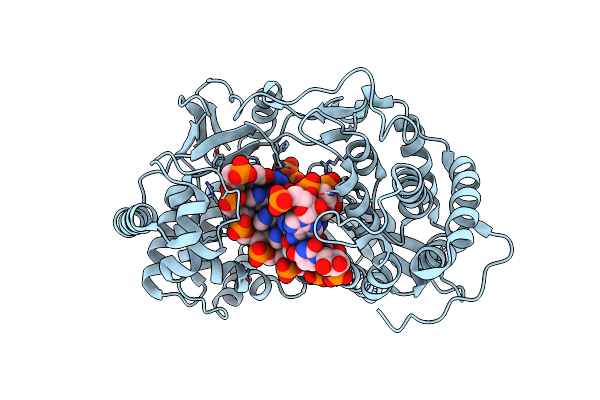
Human Pd-L1 Bound To A Macrocyclic Peptide Which Blocks The Pd-1/Pd-L1 Interaction
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-01-01 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Neta Hairpoin Loop Deletion In Complex With Gs-639476 (Diphsohate Version Of Gs-9813), Mn2+ And Symmetrical Primer Template 5'-Auaaauuu
Organism: Escherichia coli, Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-03-22 Classification: immune system/RNA Ligands: MN, 8B4, CL |
 |
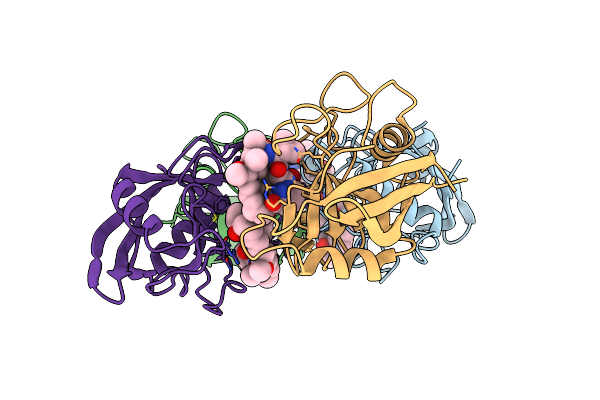
Structure Determination Of A Potent, Selective Antibody Inhibitor Of Human Mmp9 (Gs-5745 Bound To Mmp-9)
Organism: Oryctolagus cuniculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-03-15 Classification: hydrolase/hydrolase inhibitor Ligands: ZN, CA, NCO |
 |
Structure Determination Of A Potent, Selective Antibody Inhibitor Of Human Mmp9 (Apo Mmp9)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-01 Classification: hydrolase/hydrolase inhibitor Ligands: ZN, CA |
 |
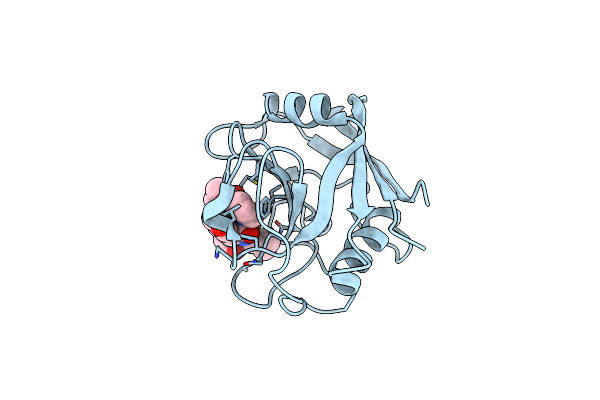
Discovery Of A Potent Cyclophilin Inhibitor (Compound 3) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 7HG |
 |
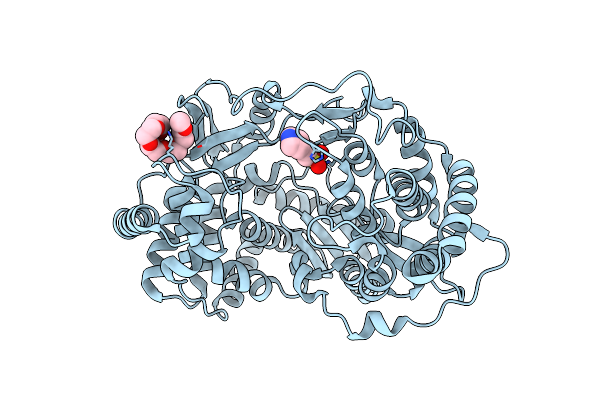
Discovery Of A Potent Cyclophilin Inhibitor (Compound 5) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 78E |
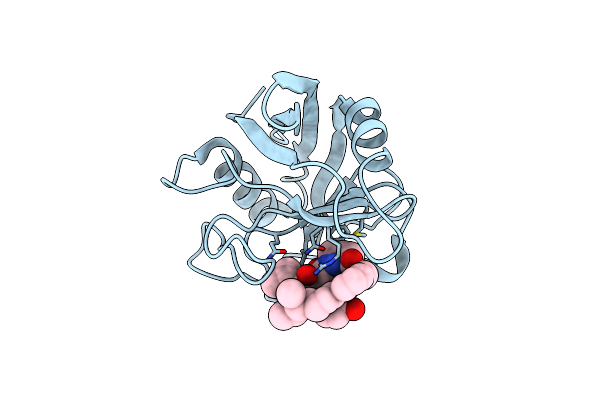 |
Discovery Of A Potent Cyclophilin Inhibitor (Compound 6) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 78R |
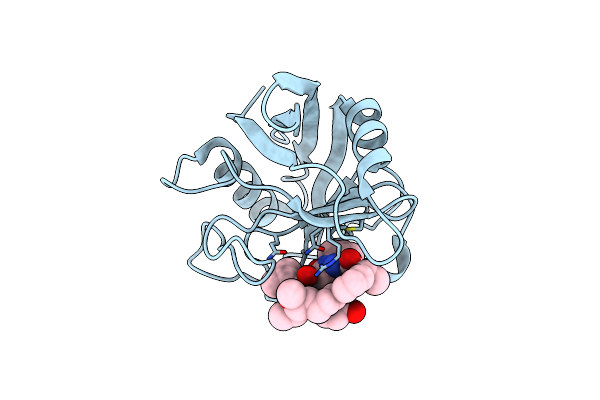 |
Discovery Of A Potent Cyclophilin Inhibitor (Compound 7) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 78X |
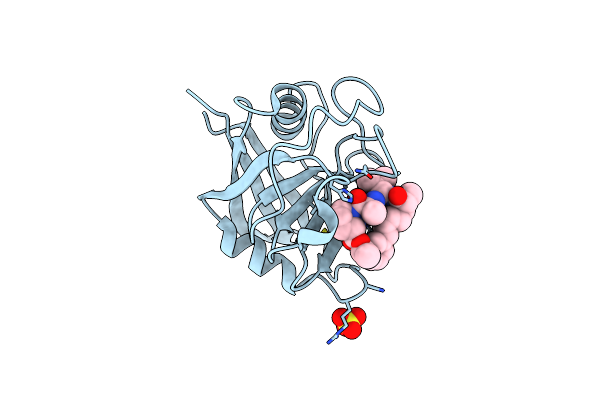 |
Discovery Of A Potent Cyclophilin Inhibitor (Compound 8) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 838, SO4 |
 |
Apo Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With E86Q E87Q S15G C223H V321I And Delta8 Mutations
Organism: Hepatitis c virus jfh-1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-02-11 Classification: TRANSFERASE Ligands: CL, EPE, PG6 |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Udp, Mn2+ And Symmetrical Primer Template 5'-Caaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, UDP |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Cdp, Mn2+ And Symmetrical Primer Template 5'-Agaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, CDP |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Adp, Mn2+ And Symmetrical Primer Template 5'-Auaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, ADP, CL |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Gdp, Mn2+ And Symmetrical Primer Template 5'-Acaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, GDP |