Search Count: 14
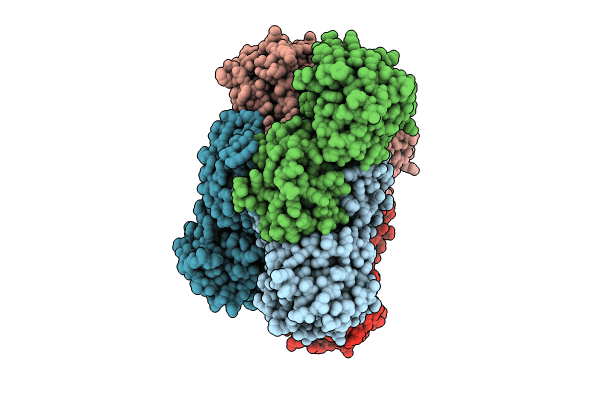 |
Crystal Structure Of Glyoxylate Reductase From Acetobacter Aceti In The Apo Form
Organism: Acetobacter aceti
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-10-23 Classification: OXIDOREDUCTASE |
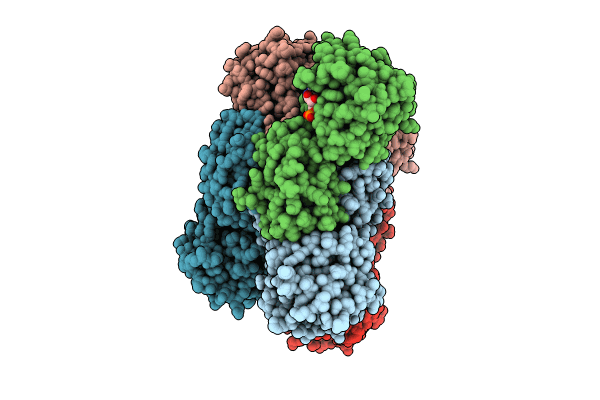 |
Crystal Structure Of Glyoxylate Reductase From Acetobacter Aceti In Complex With Nadh
Organism: Acetobacter aceti
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-10-23 Classification: OXIDOREDUCTASE Ligands: NAI |
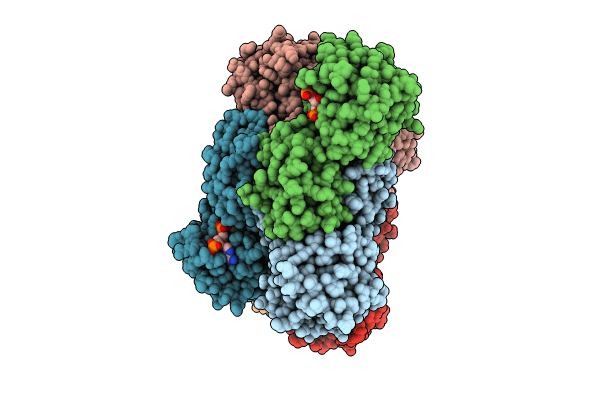 |
Crystal Structure Of Glyoxylate Reductase From Acetobacter Aceti In Complex With Nadph
Organism: Acetobacter aceti
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-10-23 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-02-04 Classification: LYASE Ligands: MG, CAP, EDO |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2013-05-15 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Amp Phosphorylase C-Terminal Deletion Mutant In The Apo-Form
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2013-05-15 Classification: TRANSFERASE |
 |
Crystal Structure Of Amp Phosphorylase C-Terminal Deletion Mutant In Complex With Substrates
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2013-05-15 Classification: TRANSFERASE Ligands: AMP, SO4 |
 |
Crystal Structure Of Ribose-1,5-Bisphosphate Isomerase From Thermococcus Kodakarensis Kod1 In Complex With Alpha-D-Ribose-1,5-Bisphosphate
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2012-04-25 Classification: ISOMERASE Ligands: RI2, PEG, CL, MG, PG4 |
 |
Crystal Structure Of Ribose-1,5-Bisphosphate Isomerase From Thermococcus Kodakaraensis Kod1 In Complex With Ribulose-1,5-Bisphosphate
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-11-03 Classification: ISOMERASE Ligands: RUB, PEG, MG |
 |
Crystal Structure Of Ribose-1,5-Bisphosphate Isomerase From Thermococcus Kodakaraensis Kod1
Organism: Thermococcus kodakaraensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-03-31 Classification: ISOMERASE Ligands: PEG, MG |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 1999-07-29 Classification: OXIDOREDUCTASE Ligands: CU, MG, NA, HEA, ZN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1999-07-22 Classification: OXIDOREDUCTASE Ligands: CU, MG, NA, HEA, CMO, ZN |
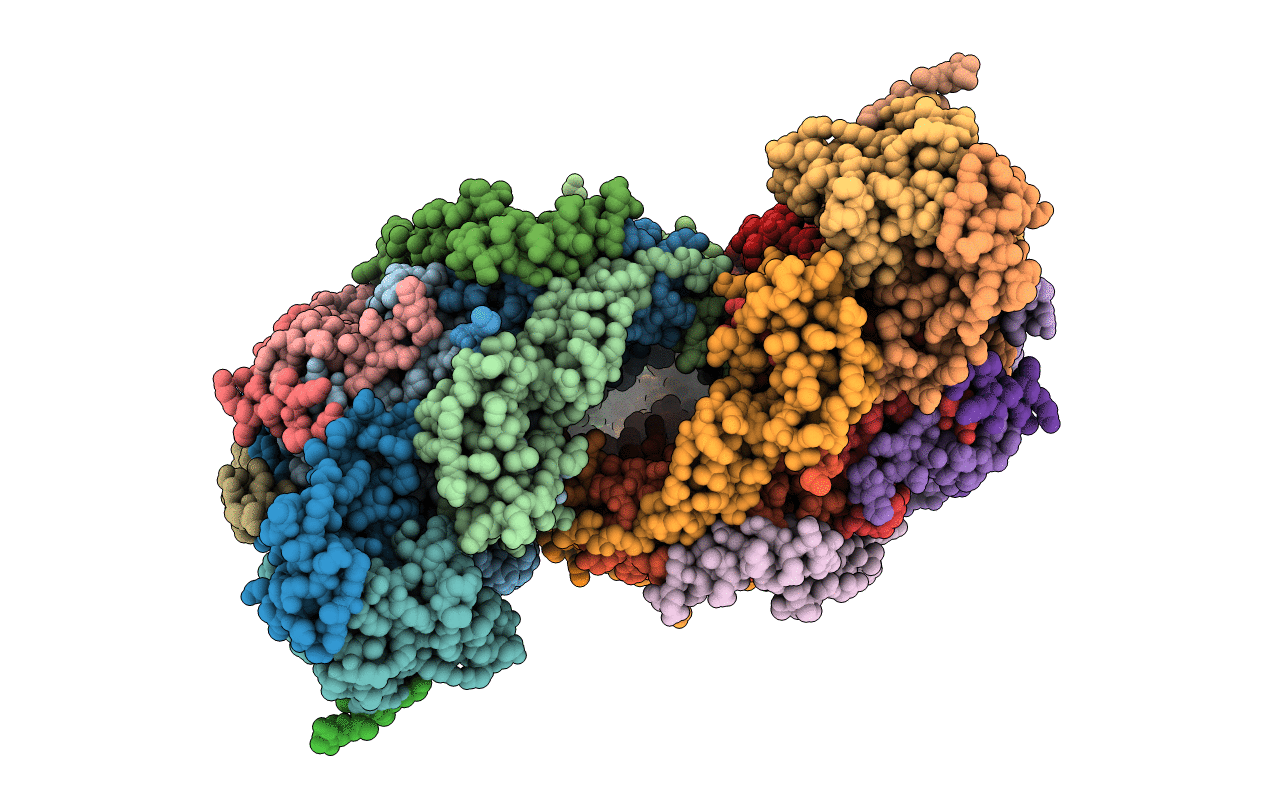 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1999-01-13 Classification: OXIDOREDUCTASE Ligands: CU, MG, NA, HEA, PER, ZN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1996-12-07 Classification: OXIDOREDUCTASE (CYTOCHROME(C)-OXYGEN) Ligands: CU, MG, HEA, ZN |

