Search Count: 23
 |
Organism: Shigella flexneri 5a str. m90t, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-05-07 Classification: CYTOSOLIC PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG |
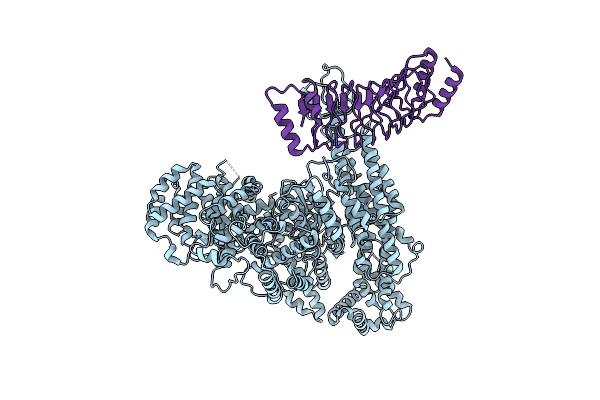 |
Cryo-Em Focused Refined Map Of Human Rnf213 E3 Module And Ipah1.4 Lrr Domain
Organism: Shigella flexneri, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: C8X |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: J1K |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: J1K |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: J1K |
 |
Crystal Structure Of A Novel Alpha/Beta Hydrolase In Apo Form In Complex With Citrate
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CIT, MPD, SO4 |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: PGE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-08-25 Classification: HYDROLASE |
 |
Organism: Unidentified, Streptomyces echinatus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2018-05-23 Classification: ANTIBIOTIC/DNA Ligands: MG, MN, QUI |
 |
Organism: Unidentified, Streptomyces echinatus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-05-23 Classification: ANTIBIOTIC/DNA Ligands: MN, QUI, K |
 |
Sugar Transporter Of Atsweet13 In Inward-Facing State With A Substrate Analog
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2017-09-13 Classification: TRANSPORT PROTEIN Ligands: DCM |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2017-08-30 Classification: DNA Ligands: CO |
 |
Crystal Structure Of Klebsiella Pneumoniae Pmra In Complex With Pmra Box Dna
Organism: Klebsiella pneumoniae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-11-11 Classification: TRANSCRIPTION/DNA Ligands: BEF, MG |
 |
Crystal Structure Of Klebsiella Pneumoniae Pmra In Complex With Pmra Box Dna
Organism: Klebsiella pneumoniae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2015-11-11 Classification: TRANSCRIPTION/DNA Ligands: BEF, MG |
 |
Structural Basis Of Dna Recognition By The Effector Domain Of Klebsiella Pneumoniae Pmra
Organism: Klebsiella pneumoniae
Method: SOLUTION NMR Release Date: 2014-01-22 Classification: SIGNALING PROTEIN |
 |
Solution Structure Of The C-Terminal Np-Repeat Domain Of Tic40, A Co-Chaperone During Protein Import Into Chloroplasts
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2012-11-14 Classification: PROTEIN TRANSPORT |
 |
Organism: Dermatophagoides farinae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-10-17 Classification: ALLERGEN |

