Search Count: 9
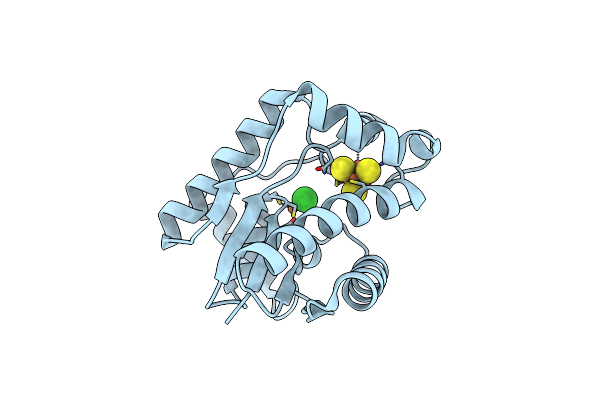 |
Crystal Structure Of Epoxyqueuosine Reductase Queh From Thermotoga Maritima
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-11-03 Classification: OXIDOREDUCTASE Ligands: SF4, FE, CL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2021-02-17 Classification: LYASE Ligands: MN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2020-09-30 Classification: LYASE Ligands: MN, GOL, CL, NA |
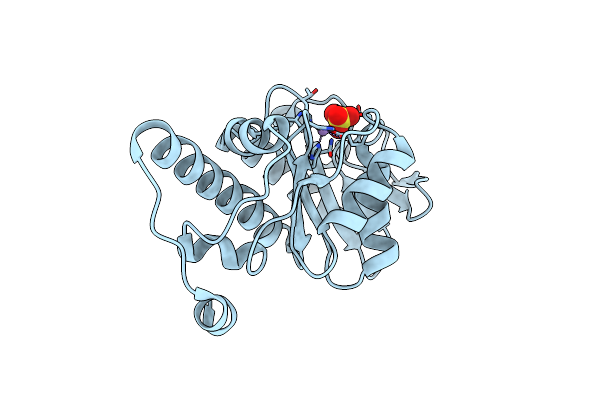 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase From Bacillus Thuringiensis Involved In Dispatching The Ubiquitous Radical Sam Enzyme Byproduct 5-Deoxyribose
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-07-04 Classification: LYASE Ligands: SO4, MN |
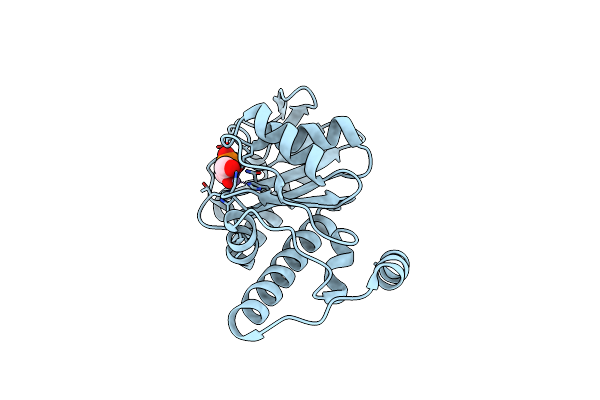 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase Bound With Dhap From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-07-04 Classification: LYASE Ligands: MN, 13P |
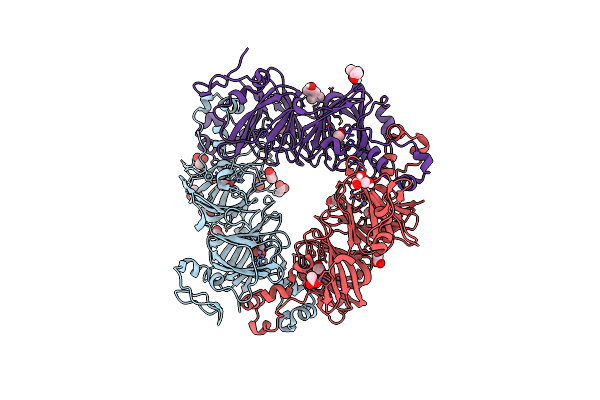 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-10-18 Classification: LYASE Ligands: ACT, GOL, MN, MPD |
 |
Assigning The Epr Fine Structure Parameters Of The Mn(Ii) Centers In Bacillus Subtilis Oxalate Decarboxylase By Site-Directed Mutagenesis And Dft/Mm Calculations
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-03-05 Classification: LYASE Ligands: CO |
 |
A Structural Element That Modulates Proton-Coupled Electron Transfer In Oxalate Decarboxylase
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2012-04-25 Classification: LYASE Ligands: MN, CO3, EDO, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1998-01-07 Classification: OXIDOREDUCTASE Ligands: MN |

