Search Count: 55
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: PC1 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: TRANSLOCASE Ligands: FUD, PC1 |
 |
Organism: Dragon grouper nervous necrosis virus
Method: SOLUTION NMR Release Date: 2024-10-30 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph8.0 (3.23A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph6.5 (2.82A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph5.0 (3.52A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virion At Ph6.5 (3.12A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-03-27 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, A1H01, IMD, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2024-03-27 Classification: CELL CYCLE Ligands: GTP, MG, CA, IMD, GDP, MES, A1H00, GOL, ACP |
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: 140 |
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: 9HO |
 |
Organism: Escherichia coli, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NFI |
 |
Structural Basis Of Transcriptional Activation By The Ompr/Phob-Family Response Regulator Pmra
Organism: Escherichia coli bl21(de3), Klebsiella pneumoniae jm45, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-05-11 Classification: HYDROLASE |
 |
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: BIOSYNTHETIC PROTEIN Ligands: NI |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2021-11-10 Classification: VIRAL PROTEIN Ligands: H3F |
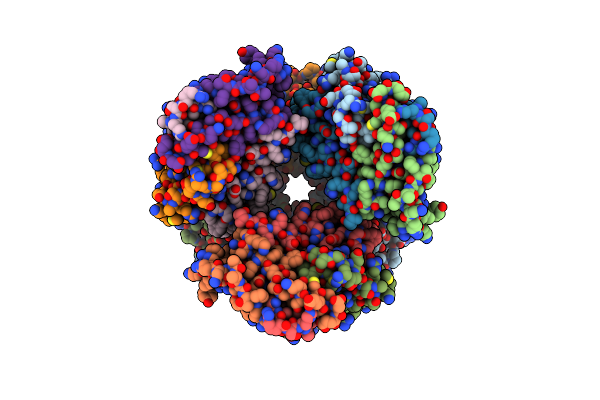 |
Cryoem Structure Of Particulate Methane Monooxygenase Associated With Cu(I)
Organism: Methylococcus capsulatus (strain atcc 33009 / ncimb 11132 / bath)
Method: ELECTRON MICROSCOPY Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: CU1 |
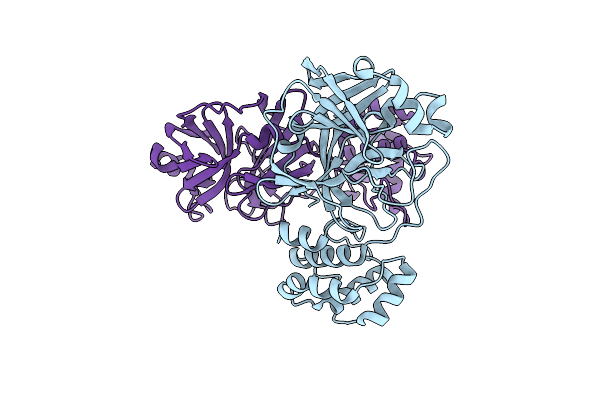 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-05-05 Classification: VIRAL PROTEIN |
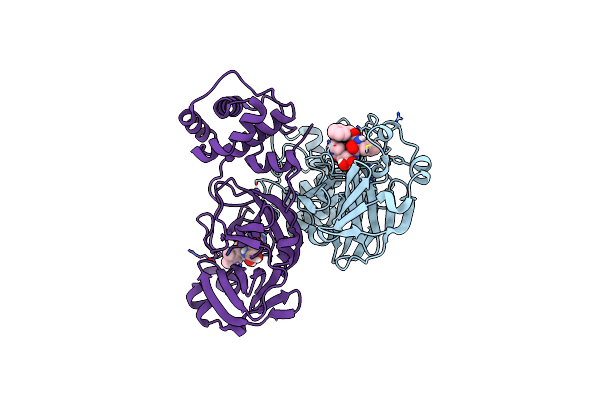 |
1.7A Resolution Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With Broad-Spectrum Coronavirus Protease Inhibitor Gc376
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2021-05-05 Classification: VIRAL PROTEIN |

