Search Count: 18
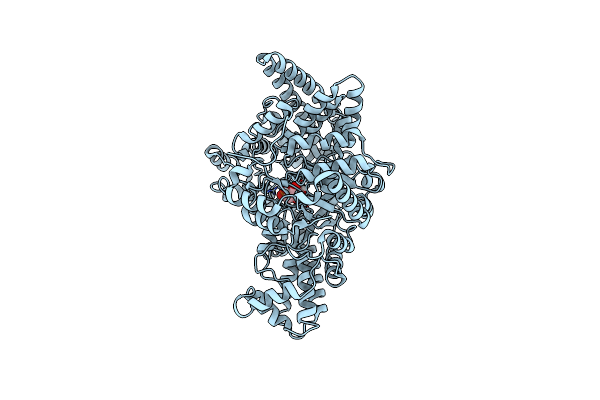 |
Crystal Structure Of Pfld Bound To 1,5-Anhydromannitol-6-Phosphate In Streptococcus Dysgalactiae Subsp. Equisimilis
Organism: Streptococcus dysgalactiae subsp. equisimilis
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 7P6 |
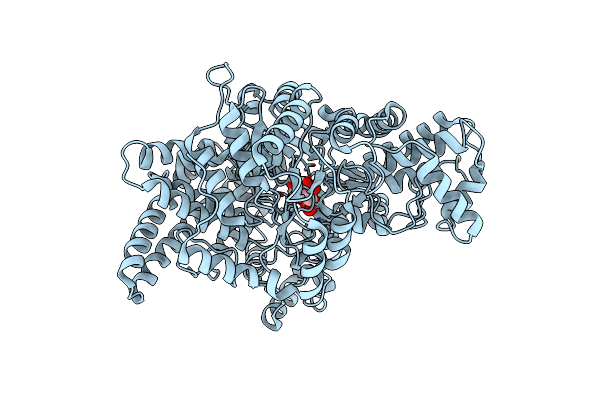 |
Crystal Structure Of Ybiw In Complex With 1,5-Anhydroglucitol-6-Phosphate In Escherichia Coli
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 0WK |
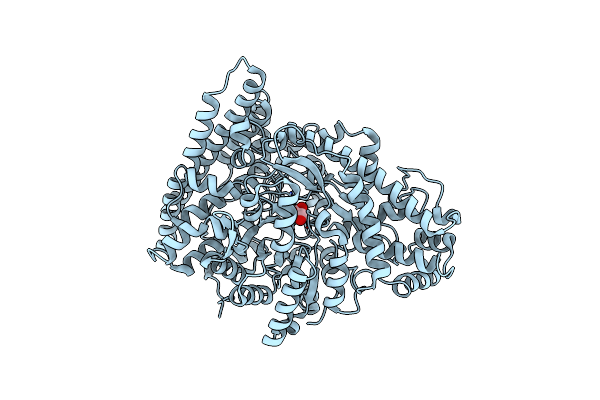 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-10-02 Classification: LYASE Ligands: GOL |
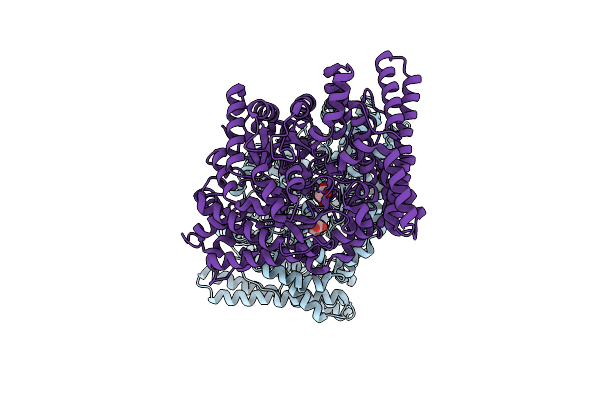 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-02 Classification: LYASE Ligands: MLI |
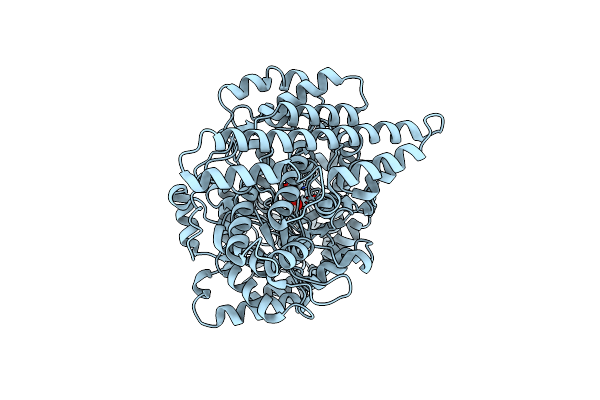 |
Crystal Structure Of N-Methyl-Cis-4-Hydroxy-L-Proline Dehydratase In Intestinibacter Bartlettii
Organism: Intestinibacter bartlettii
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2024-03-27 Classification: LYASE Ligands: 6IW |
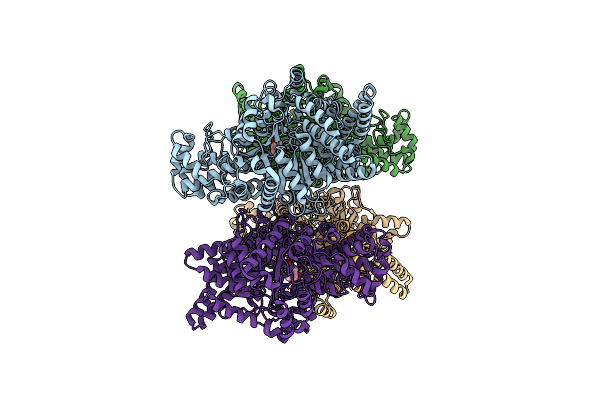 |
Crystal Structure Of N-Methyl-Cis-4-Hydroxy-D-Proline Dehydratase In Clostridium Sp. Fs41
Organism: Clostridium sp. fs41
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-03-27 Classification: LYASE Ligands: 87I |
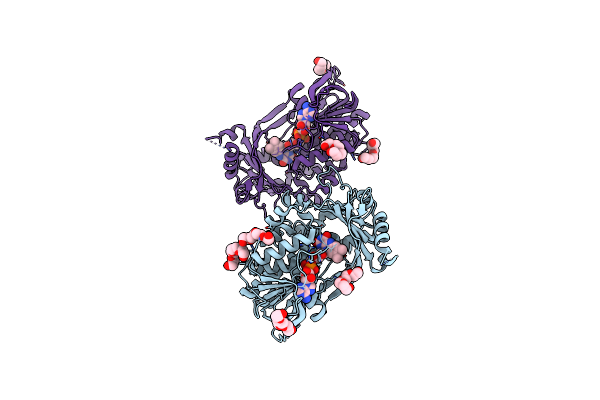 |
Organism: Floropilus chiversii
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-07-26 Classification: FLAVOPROTEIN Ligands: FAD, PG4, CL |
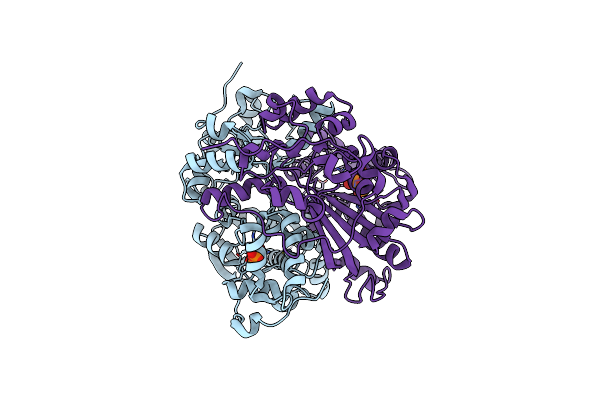 |
Organism: Rhodococcus wratislaviensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: ZN, DPO, GLY |
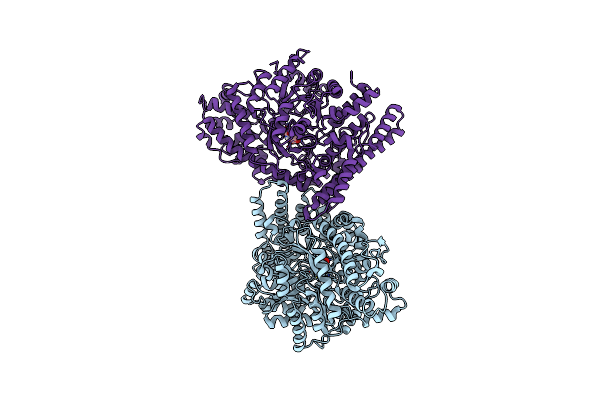 |
Anaerobic Hydroxyproline Degradation Involving C-N Cleavage By A Glycyl Radical Enzyme
Organism: Clostridiales bacterium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-06-01 Classification: LYASE Ligands: UY7 |
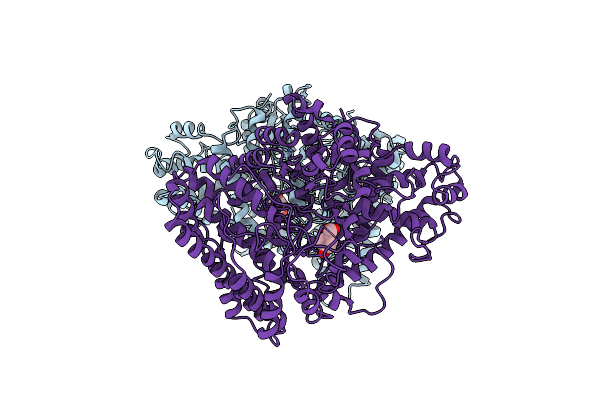 |
The Crystal Structure Of Arylacetate Decarboxylase From Olsenella Scatoligenes.
Organism: Olsenella scatoligenes
Method: X-RAY DIFFRACTION Resolution:3.53 Å Release Date: 2021-07-28 Classification: LYASE Ligands: 4HP |
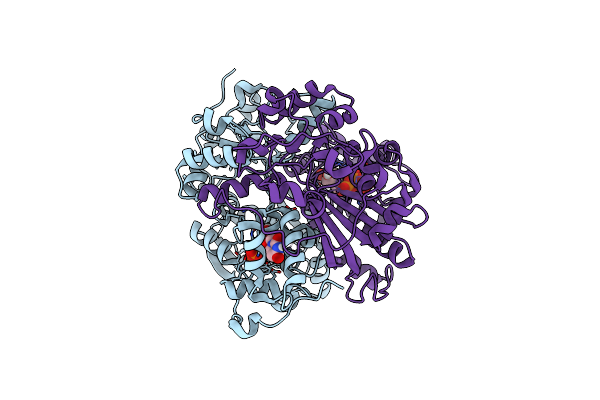 |
Organism: Rhodococcus wratislaviensis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-07-28 Classification: HYDROLASE Ligands: DUT, ZN |
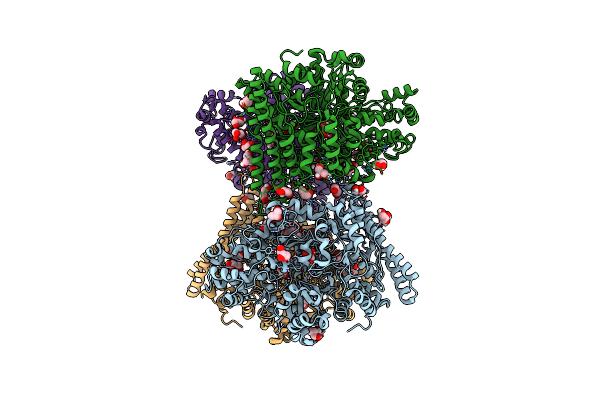 |
Organism: Bilophila wadsworthia 3_1_6
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-06-24 Classification: LYASE Ligands: LLQ, GOL, NA |
 |
The Cyrstal Structure Of Taurine:2-Oxoglutarate Aminotransferase From Bifidobacterium Kashiwanohense, In Complex With Plp And Glutamate
Organism: Bifidobacterium kashiwanohense pv20-2
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-01-01 Classification: TRANSFERASE Ligands: GLU, PLP |
 |
Crystal Structure Of Sulfoacetaldehyde Reductase From Bifidobacterium Kashiwanohense
Organism: Bifidobacterium kashiwanohense pv20-2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-12 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
Crystal Structure Of Sulfoacetaldehyde Reductase From Bifidobacterium Kashiwanohense In Complex With Nad+
Organism: Bifidobacterium kashiwanohense pv20-2
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2019-06-12 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
 |
Organism: Desulfovibrio vulgaris (strain hildenborough / atcc 29579 / dsm 644 / ncimb 8303)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-03-20 Classification: LYASE Ligands: 8X3, GOL |
 |
The Crystal Structure Of Sulfoacetaldehyde Reductase From Klebsiella Oxytoca
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-02-13 Classification: CYTOSOLIC PROTEIN Ligands: NDP, 8X3 |
 |
Organism: Streptomyces sp. ma37
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-10-26 Classification: TRANSFERASE Ligands: MET, ADN |

