Search Count: 16
 |
G13D Mutant Of Kras4B (2-169) Bound To Gdp With The Switch-I In Fully Open Conformation Crystallized In Sodium Potassium Phosphate Buffer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-06-12 Classification: ONCOPROTEIN Ligands: GDP, NA |
 |
G13D Mutant Of Kras4B (2-169) Bound To Gdp With The Switch-I In Fully Open Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-22 Classification: ONCOPROTEIN Ligands: GDP |
 |
Organism: Severe acute respiratory syndrome coronavirus, Tequatrovirus t4
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: BIOSYNTHETIC PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus, Enterobacteria phage t4 (bacteriophage t4)
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: BIOSYNTHETIC PROTEIN Ligands: NAG |
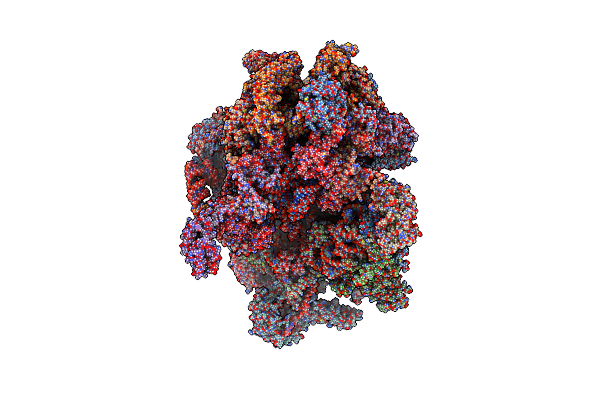 |
Human Mitochondrial Ribosome In Complex With Mrna, A/A-, P/P- And E/E-Trnas At 2.63 A Resolution
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: RIBOSOME Ligands: NAD, SPM, SPD, MG, K, ZN, FES, ATP, GDP, PUT, VAL |
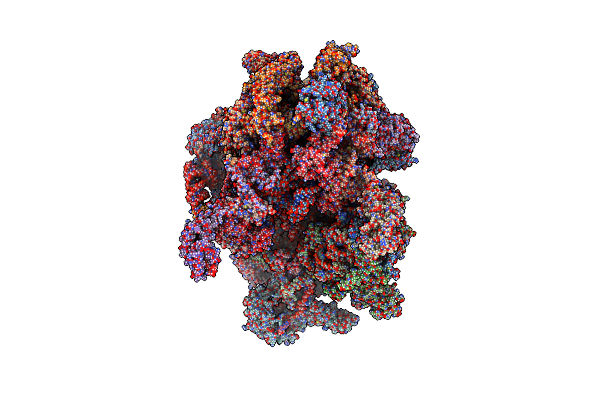 |
Human Mitochondrial Ribosome In Complex With Mrna, A/P- And P/E-Trnas At 2.98 A Resolution
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: RIBOSOME Ligands: NAD, SPM, SPD, K, MG, ZN, FES, ATP, GDP, PUT, VAL |
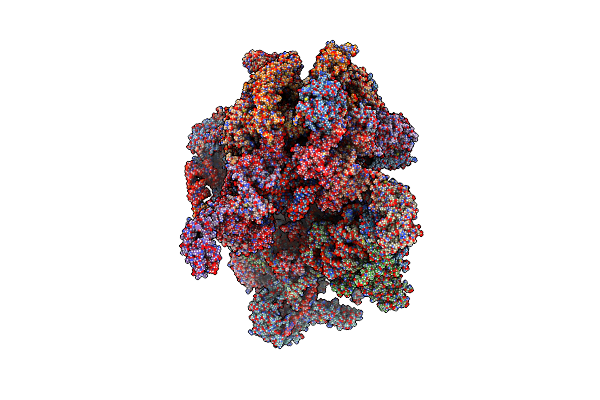 |
Human Mitochondrial Ribosome At 2.2 A Resolution (Bound To Partly Built Trnas And Mrna)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-07-13 Classification: RIBOSOME Ligands: NAD, SPM, SPD, MG, K, ZN, FES, ATP, GDP, PUT, VAL |
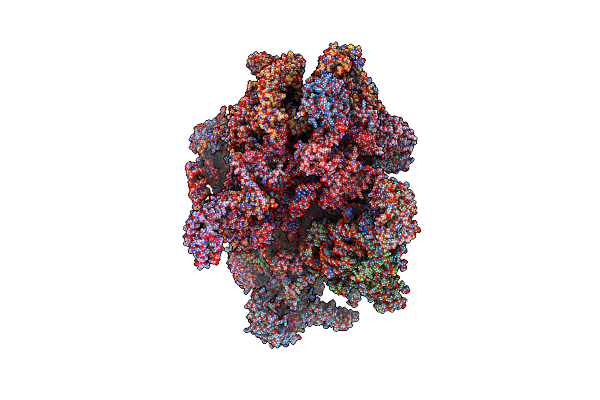 |
Human Mitochondrial Ribosome In Complex With Oxa1L, Mrna, A/A Trna, P/P Trna And Nascent Polypeptide
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-01-13 Classification: RIBOSOME Ligands: MG, K, ZN, FES, ATP, GTP, ALA |
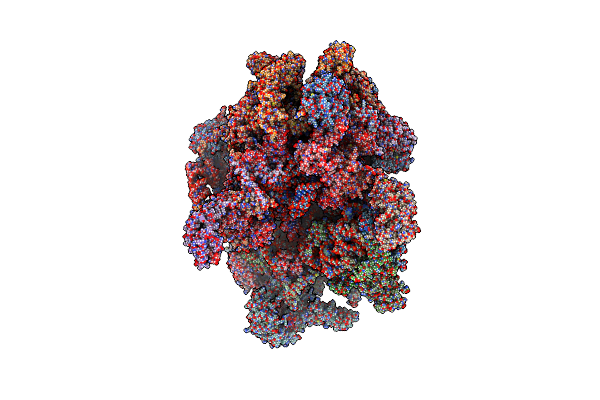 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-01-13 Classification: RIBOSOME Ligands: MG, K, ZN, FES, ATP, GTP |
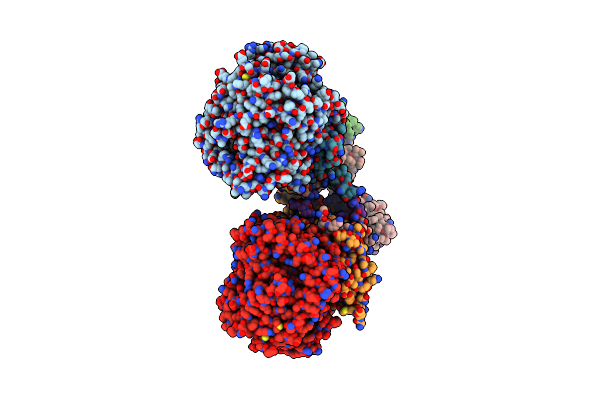 |
Crystal Structure Of Menaquinol:Oxidoreductase In Complex With Oxaloacetate
Organism: Escherichia coli 042, Escherichia coli 536, Escherichia coli dh1
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: FAD, OAA, FES, F3S, SF4 |
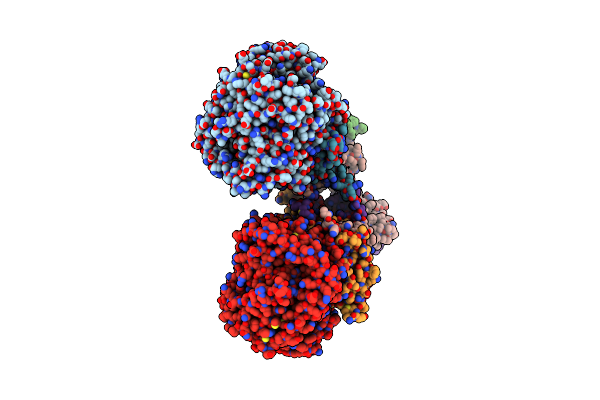 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With A 3-Nitropropionate Adduct
Organism: Escherichia coli 042, Escherichia coli o55:h7 str. cb9615
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: FAD, 3NP, FES, F3S, SF4 |
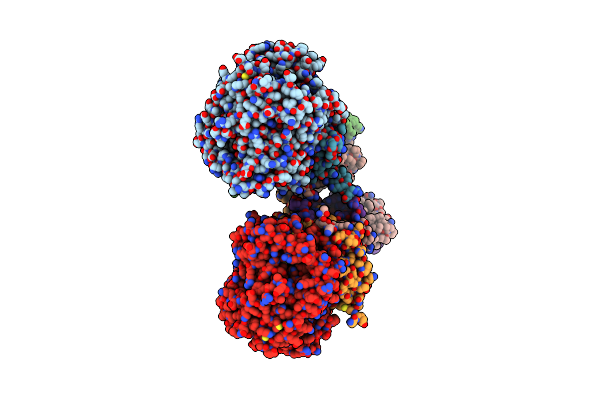 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With Fumarate
Organism: Escherichia coli 042, Escherichia coli 536, Escherichia coli dh1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-12-01 Classification: OXIDOREDUCTASE Ligands: FAD, FUM, FES, F3S, SF4 |
 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With Glutarate
Organism: Escherichia coli 042, Escherichia coli o55:h7
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2010-11-24 Classification: OXIDOREDUCTASE Ligands: FAD, GUA, FES, F3S, SF4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-03-31 Classification: LYASE Ligands: PLP |
 |
Thioredoxin A Active Site Mutants Form Mixed Disulfide Dimers That Resemble Enzyme-Substrate Reaction Intermediate
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-03-10 Classification: ELECTRON TRANSPORT Ligands: PEG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2006-02-21 Classification: OXIDOREDUCTASE Ligands: FLC, FAD, FES, F3S, SF4, MQ7 |

