Search Count: 38
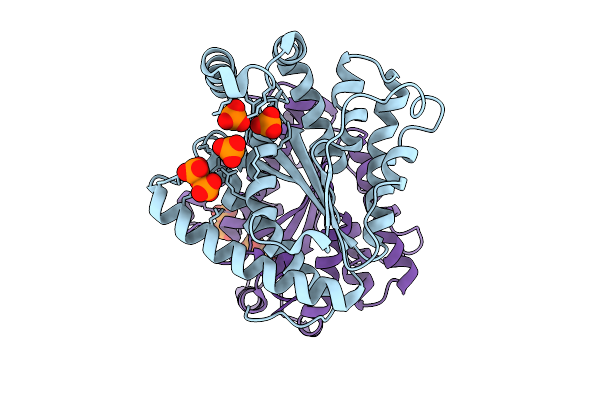 |
Crystal Structure Of Polyphosphate Kinase 2-Ii (Ppk2-Ii) From Bacillus Cereus Apo-Form
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: POP, PO4 |
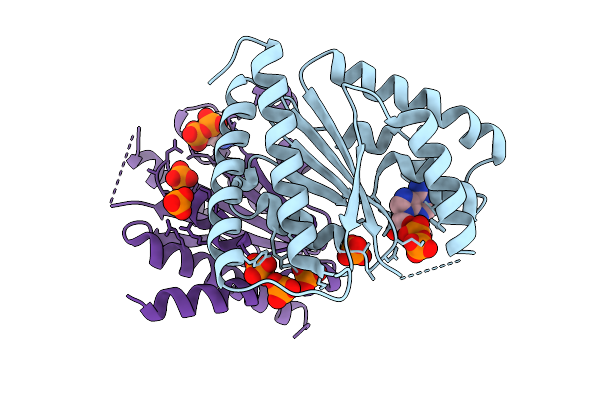 |
Crystal Structure Of Polyphosphate Kinase 2-Ii (Ppk2-Ii) From Lysinibacillus Fusiformis Bound To Adp (Form I)
Organism: Lysinibacillus fusiformis
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: ADP, PO4 |
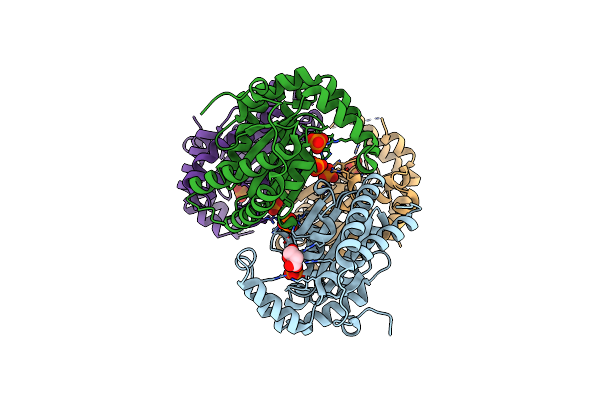 |
Crystal Structure Of Polyphosphate Kinase 2-Ii (Ppk2-Ii) From Lysinibacillus Fusiformis In Apo Form
Organism: Lysinibacillus fusiformis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: PO4, SIN |
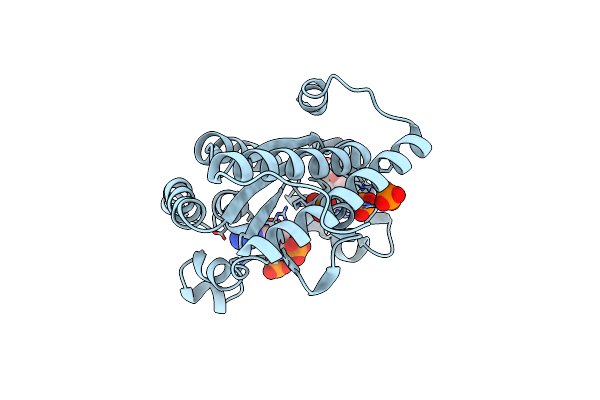 |
Crystal Structure Of Polyphosphate Kinase 2-Ii (Ppk2-Ii) From Lysinibacillus Fusiformis Bound To Amp
Organism: Lysinibacillus fusiformis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: CL, AMP, PO4 |
 |
Crystal Structure Of Polyphosphate Kinase 2-Ii (Ppk2-Ii) From Lysinibacillus Fusiformis Bound To Tmp
Organism: Lysinibacillus fusiformis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: TMP, PO4 |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase Treated At 368 K From Pyrococcus Furiosus In Complex With Inosine
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-08-21 Classification: HYDROLASE Ligands: NOS, NAD |
 |
Crystal Structure Of Sgvm Methyltransferase In Complex With Alpha-Ketoleucine And Zn2+ Ion
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: COI, CL, MG, ZN |
 |
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: ZN, COI, CL, SAM |
 |
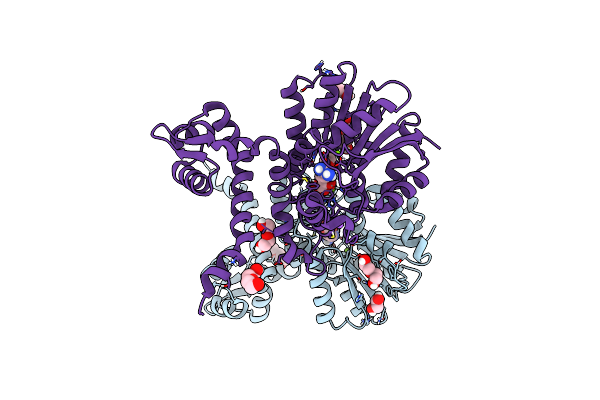
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Native-Form
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG |
 |
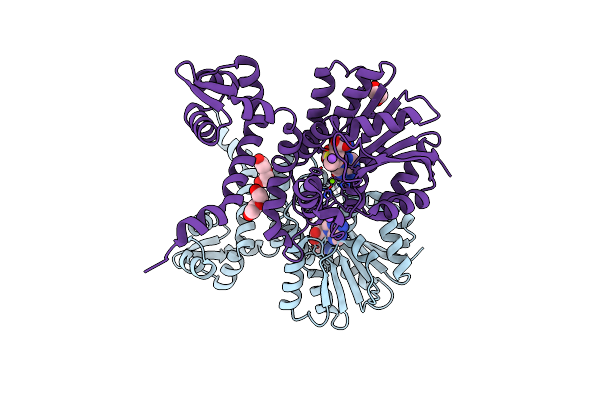
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Bound To Ketoarginine
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: NWG, MG, PEG, PGE, EDO, NA |
 |
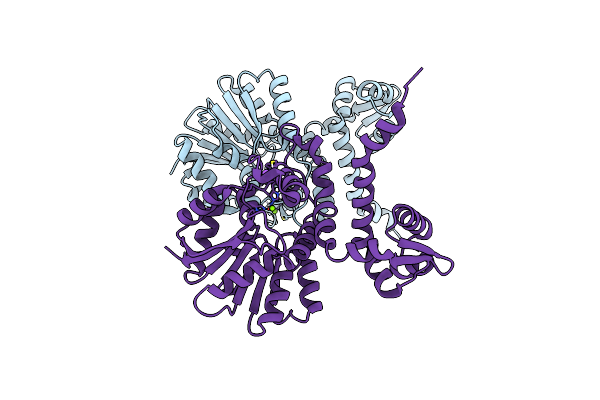
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Bound To Sam
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG, SAM, NA, PEG, EDO |
 |
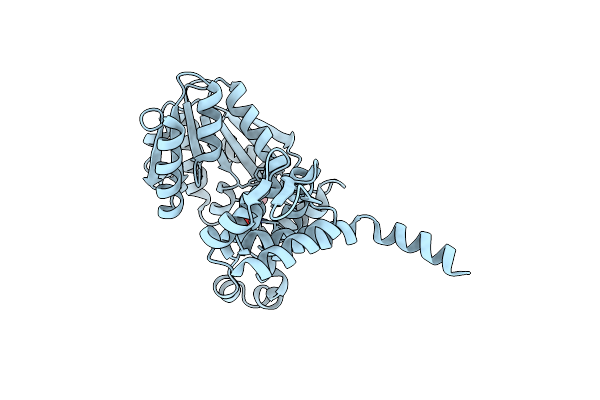
Crystal Structure Of Selenomethionine Derivatized Alpha Keto Acid C-Methyl-Transferases Mrsa
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG, NA |
 |
Crystal Structure Of Native Alpha-Keto C-Methyl Transferase Sgvm Bound To Ketoleucine
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: ZN, COI, CL |
 |
Crystal Structure Of S-Sad Phased Alpha-Keto C-Methyl Transferase Sgvm Bound To Ketoleucine
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: ZN, COI, CL, GOL |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Mus Musculus In Complex With Inosine
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NAD, NOS, NA |
 |
Crystal Structure Of O-Acetyl-L-Homoserine Sulfhydrolase From Saccharomyces Cerevisiae In Complex With Pyridoxal-5'-Phosphate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-06-21 Classification: LYASE Ligands: PLP, PEG |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Pyrococcus Furiosus In Complex With Inosine
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: NAD, NOS |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Pyrococcus Furiosus In Complex With S-Inosyl-L-Homocysteine
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: SIB, NAD |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Sulfolobus Acidocaldarius In Complex With Adenosine
Organism: Sulfolobus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: NAD, ADN |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Methanococcus Maripaludis In Complex With Inosine
Organism: Methanococcus maripaludis
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: NOS, NAD, PG4 |

