Search Count: 33
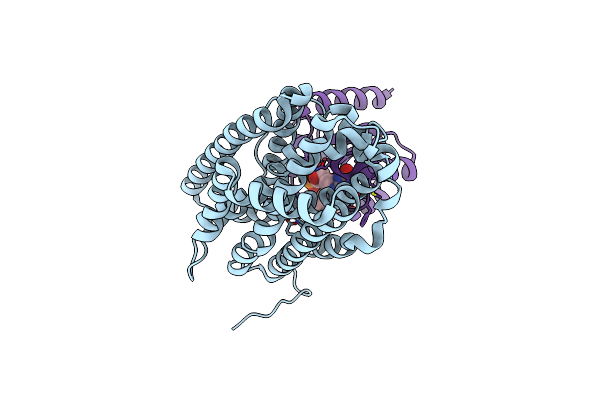 |
Crystal Structure Of Bacillus Cereus Ribonucleotide Reductase Di- Iron Nrdf In Complex With Nrdi (1.8 A Resolution)
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2014-03-19 Classification: OXIDOREDUCTASE Ligands: FE2, CL, FMN |
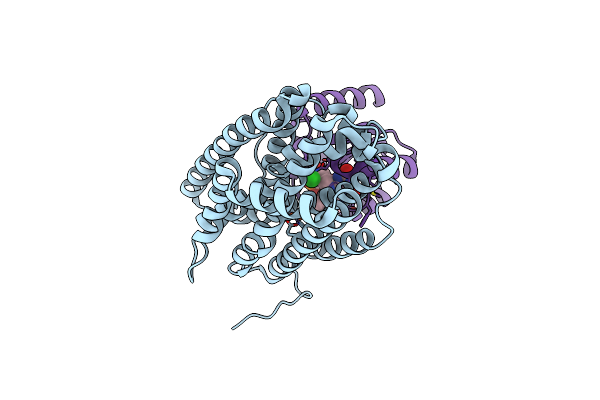 |
Crystal Structure Of Bacillus Cereus Ribonucleotide Reductase Di- Iron Nrdf In Complex With Nrdi (2.1 A Resolution)
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-03-19 Classification: OXIDOREDUCTASE Ligands: FE2, CL, FMN |
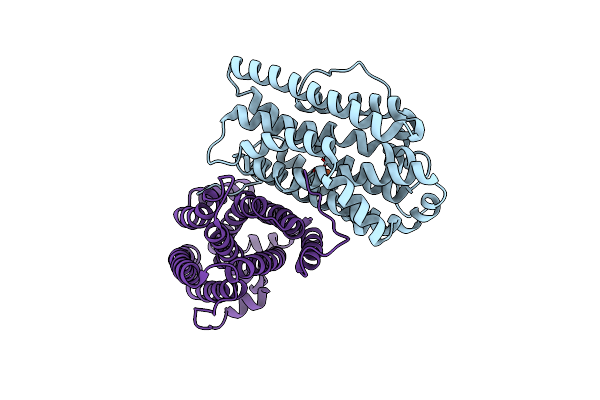 |
Crystal Structure Of Ribonucleotide Reductase Apo-Nrdf From Bacillus Cereus (Space Group C2)
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-03-19 Classification: OXIDOREDUCTASE Ligands: FE2 |
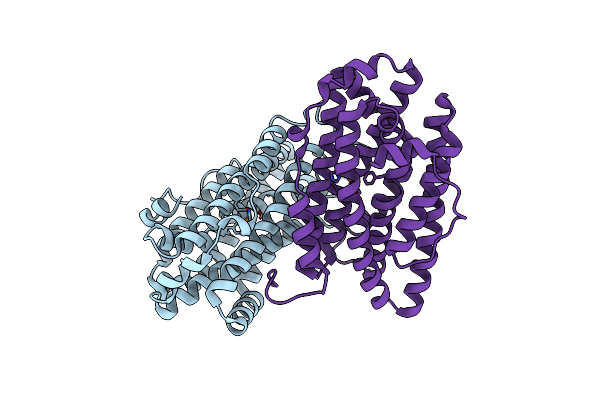 |
Crystal Structure Of Ribonucleotide Reductase Apo-Nrdf From Bacillus Cereus (Space Group P21)
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-03-19 Classification: OXIDOREDUCTASE Ligands: FE2 |
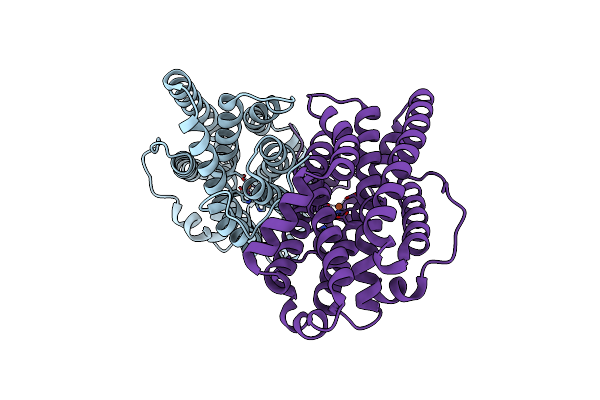 |
Crystal Structure Of Ribonucleotide Reductase Di-Iron Nrdf From Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-03-19 Classification: OXIDOREDUCTASE Ligands: FE2 |
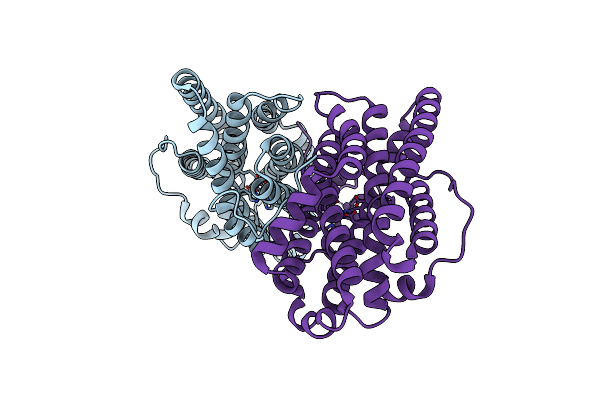 |
Crystal Structure Of Ribonucleotide Reductase Di-Manganese(Ii) Nrdf From Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-03-19 Classification: OXIDOREDUCTASE Ligands: MN |
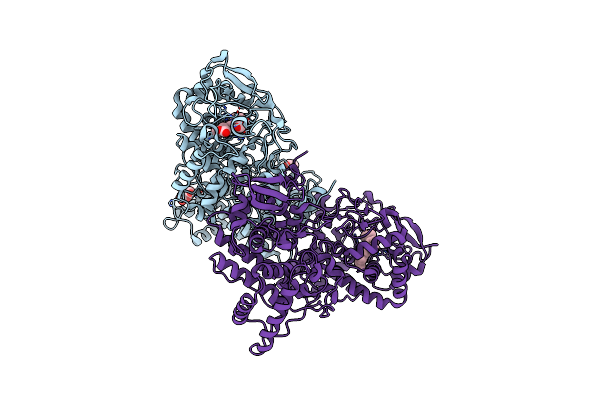 |
Crystal Structure Of The Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-11-13 Classification: OXIDOREDUCTASE Ligands: GLC, HEM, ACT |
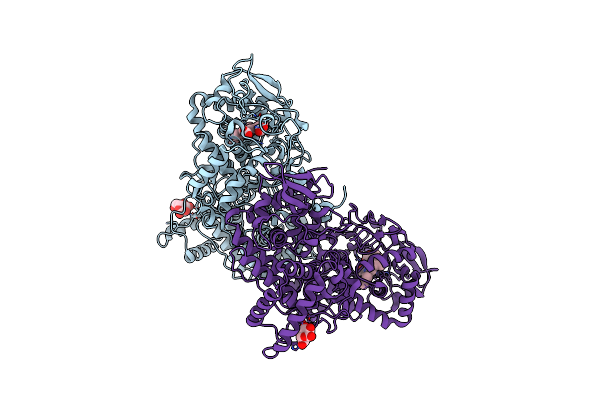 |
Crystal Structure Of The Catalase-Peroxidase (Katg) R418L Mutant From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-11-13 Classification: OXIDOREDUCTASE Ligands: HEM, GLC |
 |
Organism: Nitrosomonas europaea
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-08-14 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Crystal Structure Of N64Del Mutant Of Nitrosomonas Europaea Cytochrome C552 (Monoclinic Space Group)
Organism: Nitrosomonas europaea
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-08-14 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Crystal Structure Of N64Del Mutant Of Nitrosomonas Europaea Cytochrome C552 (Hexagonal Space Group)
Organism: Nitrosomonas europaea
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-14 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Nitrosomonas europaea atcc 19718
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2013-08-14 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-01-23 Classification: OXIDOREDUCTASE |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2013-01-23 Classification: OXIDOREDUCTASE |
 |
The Flavoprotein Nrdi From Bacillus Cereus With The Initially Oxidized Fmn Cofactor In An Intermediate Radiation Reduced State
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2010-03-09 Classification: FLAVOPROTEIN Ligands: FMN, ZN, CAC |
 |
The Flavoprotein Nrdi From Bacillus Cereus With The Initially Semiquinone Fmn Cofactor In An Intermediate Radiation Reduced State
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2010-03-09 Classification: FLAVOPROTEIN Ligands: FMN, ZN, CAC |
 |
Crystal Structure Of Peroxymyoglobin Generated By Cryoradiolytic Reduction Of Myoglobin Compound Iii
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2009-02-10 Classification: OXYGEN TRANSPORT Ligands: HEM, PER, SO4, GOL, PEO |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-01-29 Classification: OXYGEN TRANSPORT Ligands: HEM, OXY, SO4, GOL, PEO |
 |
Crystal Structure Of Peroxymyoglobin Generated By Cryoradiolytic Reduction Of Myoglobin Compound Iii
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-01-29 Classification: OXYGEN TRANSPORT Ligands: HEM, PER, SO4, GOL, PEO |
 |
Crystal Structure Of Radiation-Induced Myoglobin Compound Ii Generated After Annealing Of Peroxymyoglobin
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-01-29 Classification: OXYGEN TRANSPORT Ligands: HEM, OH, SO4, GOL, PEO |

