Search Count: 74
 |
Crystal Structure Of Light-Activated Dna-Binding Protein El222 From Erythrobacter Litoralis Crystallized And Measured In Dark.
Organism: Erythrobacter litoralis htcc2594
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-07-05 Classification: TRANSCRIPTION Ligands: FMN, CL, MG, MES |
 |
Crystal Structure Of Light-Activated Dna-Binding Protein El222 From Erythrobacter Litoralis Crystallized In Dark, Measured Illuminated.
Organism: Erythrobacter litoralis htcc2594
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-07-05 Classification: TRANSCRIPTION Ligands: FMN, CL, MG, MES |
 |
Organism: Griffithsia monilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-06-14 Classification: PHOTOSYNTHESIS Ligands: MG, CAP, EDO, BCT |
 |
Organism: Streptomyces clavuligerus atcc 27064
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-12-18 Classification: OXIDOREDUCTASE Ligands: SO4, GOL, ACT |
 |
Piperideine-6-Carboxylate Dehydrogenase From Streptomyces Clavuligerus Complexed With Nad+
Organism: Streptomyces clavuligerus atcc 27064
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-12-18 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, GOL, ACT |
 |
Piperideine-6-Carboxylate Dehydrogenase From Streptomyces Clavuligerus Complexed With Picolinic Acid
Organism: Streptomyces clavuligerus atcc 27064
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-12-18 Classification: OXIDOREDUCTASE Ligands: 6PC, SO4, GOL |
 |
Piperideine-6-Carboxylate Dehydrogenase From Streptomyces Clavuligerus Complexed With Alpha-Aminoadipic Acid
Organism: Streptomyces clavuligerus atcc 27064
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-12-18 Classification: OXIDOREDUCTASE Ligands: UN1, SO4, GOL, ACT |
 |
Unusual Posttranslational Modifications Revealed In Crystal Structures Of Diatom Rubisco.
Organism: Chaetoceros socialis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-06-27 Classification: LYASE Ligands: MG, CAP, EDO |
 |
Organism: Skeletonema marinoi
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-06-27 Classification: LYASE Ligands: MG, CAP, EDO |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2018-05-09 Classification: HYDROLASE Ligands: MRD, GOL, MPD |
 |
Organism: Thalassiosira hyalina
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-03-14 Classification: PHOTOSYNTHESIS Ligands: MG, CAP, EDO |
 |
Organism: Thalassiosira antarctica var. borealis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-02-14 Classification: PHOTOSYNTHESIS Ligands: MG, CAP, EDO |
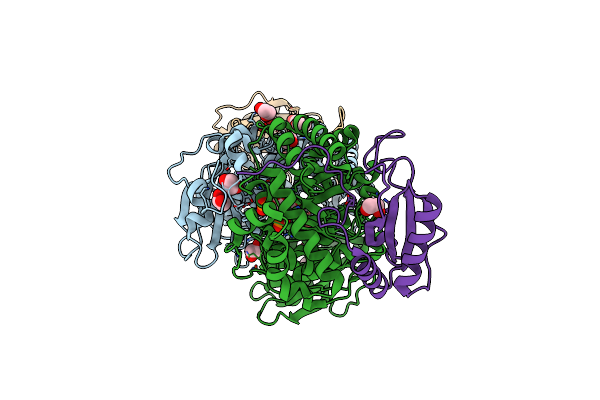 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-07-12 Classification: LYASE Ligands: MG, CAP, EDO |
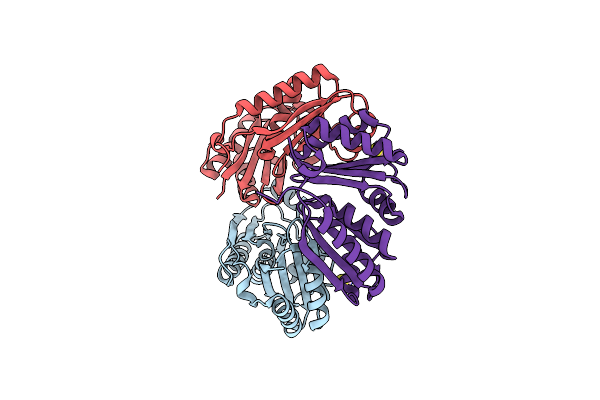 |
Organism: Synechococcus elongatus pcc 7942
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-04-12 Classification: TRANSPORT PROTEIN Ligands: SCN |
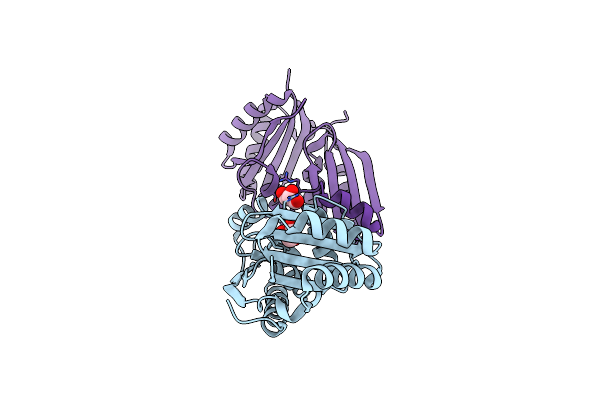 |
Organism: Synechococcus elongatus pcc 7942
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-04-12 Classification: TRANSPORT PROTEIN Ligands: GOL, CL |
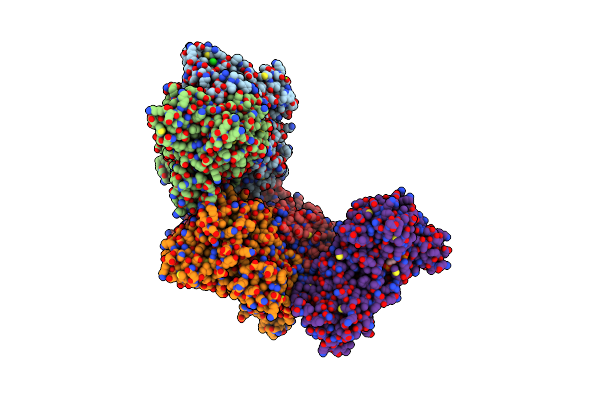 |
Crystal Structure Of Decameric Methanococcoides Burtonii Rubisco Complexed With 2-Carboxyarabinitol Bisphosphate
Organism: Methanococcoides burtonii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-02-08 Classification: LYASE Ligands: CAP, MG, CL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-04-08 Classification: CHAPERONE Ligands: SO4 |
 |
Crystal Structure Of Glycine Decarboxylase P-Protein From Synechocystis Sp. Pcc 6803, Apo Form
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: TRS, CL |
 |
Crystal Structure Of Synechocystis Sp. Pcc 6803 Glycine Decarboxylase (P-Protein), Holo Form With Pyridoxal-5'-Phosphate And Glycine
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: GLY, BCT, PO4, EDO, BCN |
 |
Crystal Structure Of Synechocystis Sp. Pcc 6803 Glycine Decarboxylase (P-Protein), Holo Form With Pyridoxal-5'-Phosphate And Glycine, Closed Flexible Loop
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: GLY, BCT, PO4, EDO |

