Search Count: 20
 |
Organism: Apilactobacillus kunkeei
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: UNKNOWN FUNCTION |
 |
Organism: Apilactobacillus kunkeei
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: UNKNOWN FUNCTION |
 |
Organism: Apilactobacillus kunkeei
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: UNKNOWN FUNCTION |
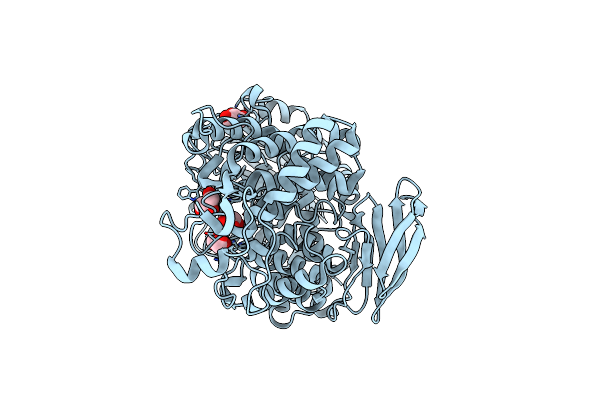 |
Crystal Structure Of Unliganded Form Of Fucob, A Gh95 Family Alpha-1,2-Fucosidase From Akkermansia Muciniphila
Organism: Akkermansia muciniphila atcc baa-835
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: GOL |
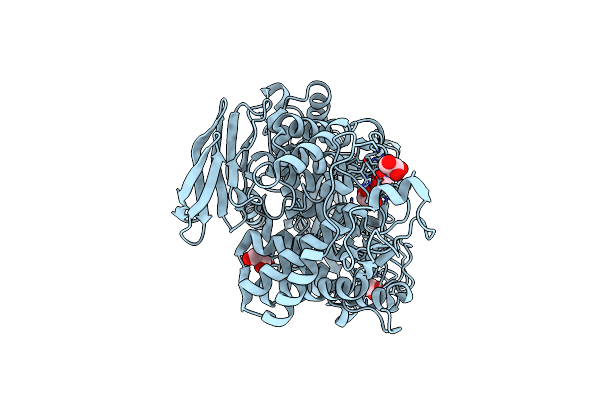 |
Crystal Structure Of Catalytic Inactive Unliganded Form Of Fucob, A Gh95 Family Alpha-1,2-Fucosidase From Akkermansia Muciniphila
Organism: Akkermansia muciniphila atcc baa-835
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: GOL |
 |
Structure And Folding Of A 600-Million-Year-Old Nuclear Coactivator Binding Domain Suggest Conservation Of Dynamic Properties
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2022-04-20 Classification: DNA BINDING PROTEIN |
 |
Structure And Folding Of A 600-Million-Year-Old Nuclear Coactivator Binding Domain Suggest Conservation Of Dynamic Properties
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2022-04-20 Classification: DNA BINDING PROTEIN |
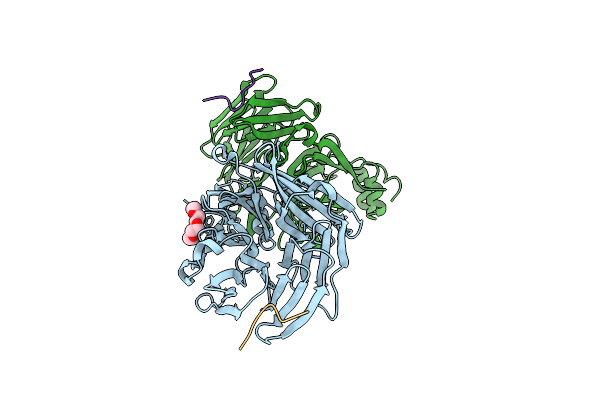 |
Clathrin Heavy Chain N-Terminal Domain Complexed With Peptide From Protein Mu-Ns Of Reovirus Type 1
Organism: Homo sapiens, Mammalian orthoreovirus 1 lang
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-03-02 Classification: TRANSPORT PROTEIN Ligands: PG4 |
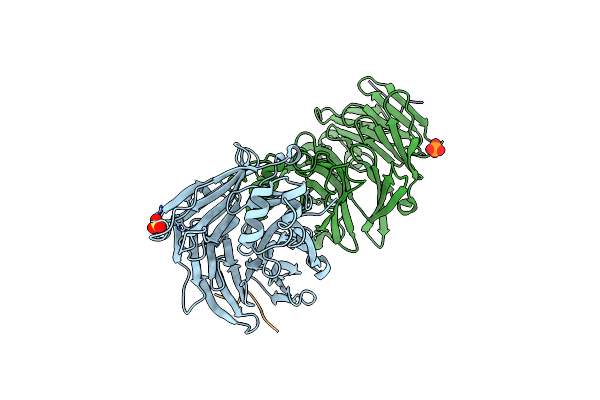 |
Clathrin Heavy Chain N-Terminal Domain Bound To Non Structured Protein 3 From Eastern Equine Encephalitis Virus
Organism: Homo sapiens, Eastern equine encephalitis virus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-03-02 Classification: TRANSPORT PROTEIN Ligands: SO4, PO4 |
 |
Crystal Structure Of C-Terminal Domain Of Pabpc1 In Complex With Nucleoprotein From Human Coronavirus 229E
Organism: Homo sapiens, Human coronavirus 229e
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-03-02 Classification: TRANSCRIPTION Ligands: SO4, GOL |
 |
Structure And Dynamics Conspire In The Evolution Of Affinity Between Intrinsically Disordered Proteins
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-10-31 Classification: DNA BINDING PROTEIN |
 |
Structure And Dynamics Conspire In The Evolution Of Affinity Between Intrinsically Disordered Proteins
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-10-31 Classification: DNA BINDING PROTEIN |
 |
Structure And Dynamics Conspire In The Evolution Of Affinity Between Intrinsically Disordered Proteins
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-10-31 Classification: DNA BINDING PROTEIN |
 |
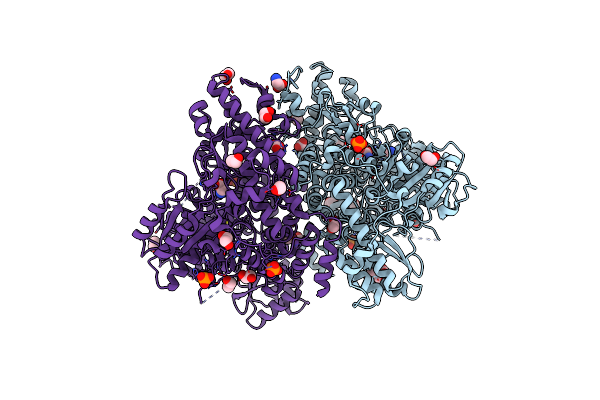
Crystal Structure Of Glycine Decarboxylase P-Protein From Synechocystis Sp. Pcc 6803, Apo Form
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: TRS, CL |
 |
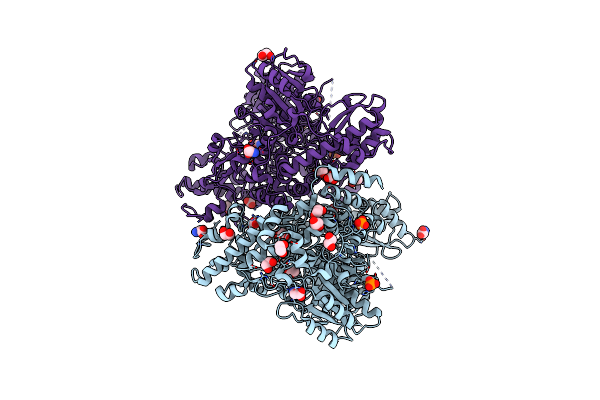
Crystal Structure Of Synechocystis Sp. Pcc 6803 Glycine Decarboxylase (P-Protein), Holo Form With Pyridoxal-5'-Phosphate And Glycine
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: GLY, BCT, PO4, EDO, BCN |
 |
Crystal Structure Of Synechocystis Sp. Pcc 6803 Glycine Decarboxylase (P-Protein), Holo Form With Pyridoxal-5'-Phosphate And Glycine, Closed Flexible Loop
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: GLY, BCT, PO4, EDO |
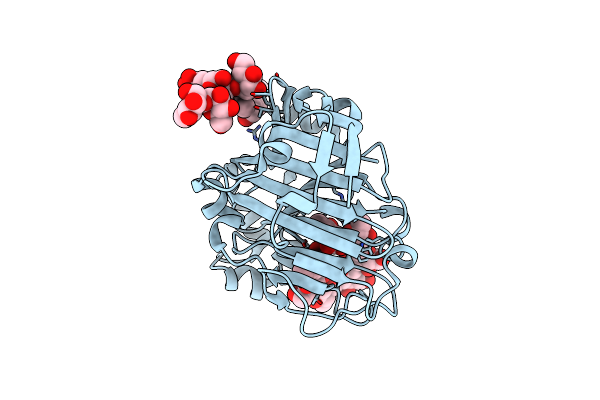 |
Nucleophile-Disabled Lam16A Mutant Holds Laminariheptaose (L7) In A Cyclical Conformation
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-01-26 Classification: HYDROLASE |
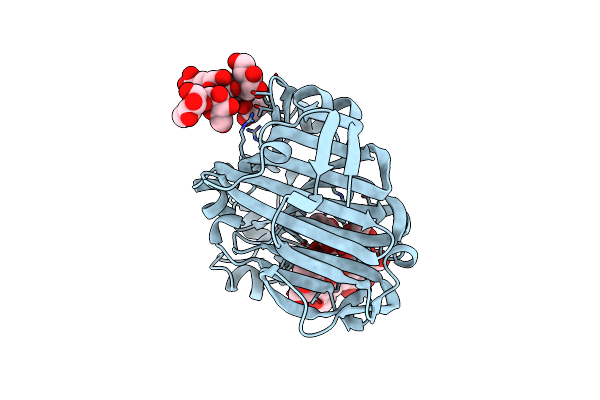 |
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2010-01-26 Classification: HYDROLASE |
 |
Glc(Beta-1-3)Glc Disaccharide In -1 And -2 Sites Of Laminarinase 16A From Phanerochaete Chrysosporium
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2009-07-21 Classification: HYDROLASE Ligands: NAG, LGC |
 |
2 Beta-Glucans (6-O-Glucosyl-Laminaritriose) In Both Donor And Acceptor Sites Of Gh16 Laminarinase 16A From Phanerochaete Chrysosporium.
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2009-07-21 Classification: HYDROLASE |

