Search Count: 22
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-02-24 Classification: LYASE Ligands: PO4, NA, PRO, MTA |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Release Date: 2021-02-24 Classification: LYASE Ligands: PO4, SAH |
 |
Organism: Bacteriophage sp.
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-02-24 Classification: LYASE Ligands: PO4 |
 |
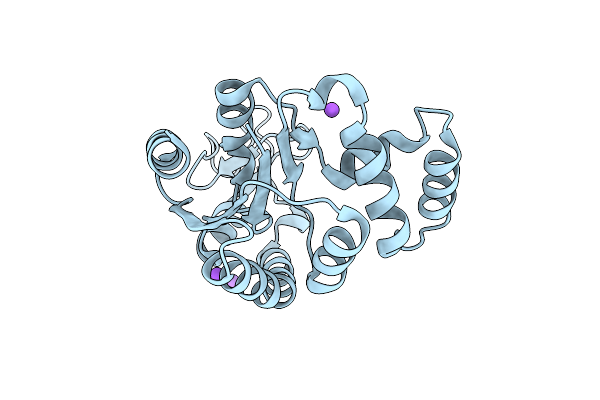 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2019-08-07 Classification: TRANSFERASE |
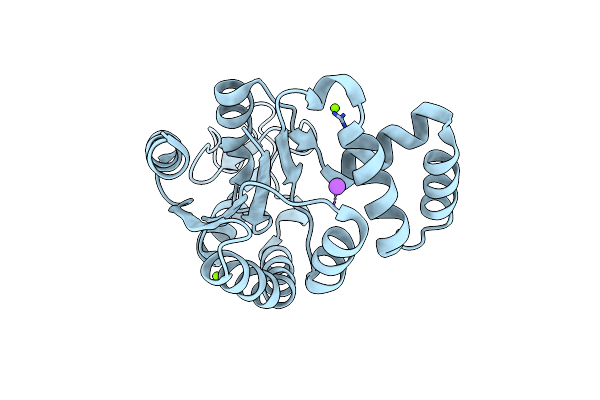 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-08-07 Classification: TRANSFERASE Ligands: CL, NA, MG |
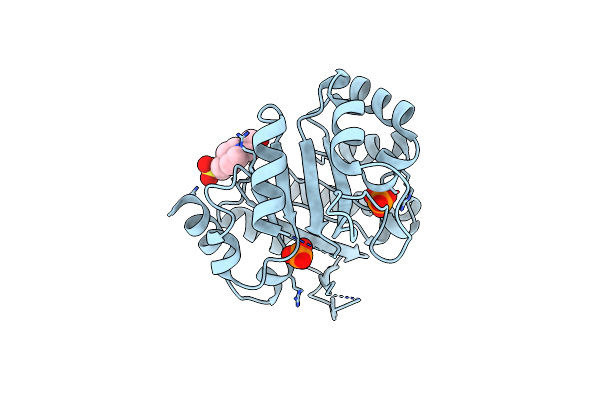 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: PO4, EPE |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: SO4, GOL, NA |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: PO4, NA |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: SO4, NA, ACY, GOL |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: GUO, GOL |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: PO4, EPE |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Salmonella choleraesuis
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2017-04-19 Classification: ISOMERASE |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-04-12 Classification: ISOMERASE Ligands: SO4, GOL |
 |
Organism: Salmonella enterica subsp. enterica serovar cubana str. 76814
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-28 Classification: ISOMERASE Ligands: 2ER, NA |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-09-28 Classification: ISOMERASE Ligands: GUO |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2016-09-28 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-09-28 Classification: ISOMERASE Ligands: GOL, NA, SO4 |

