Search Count: 1,146
 |
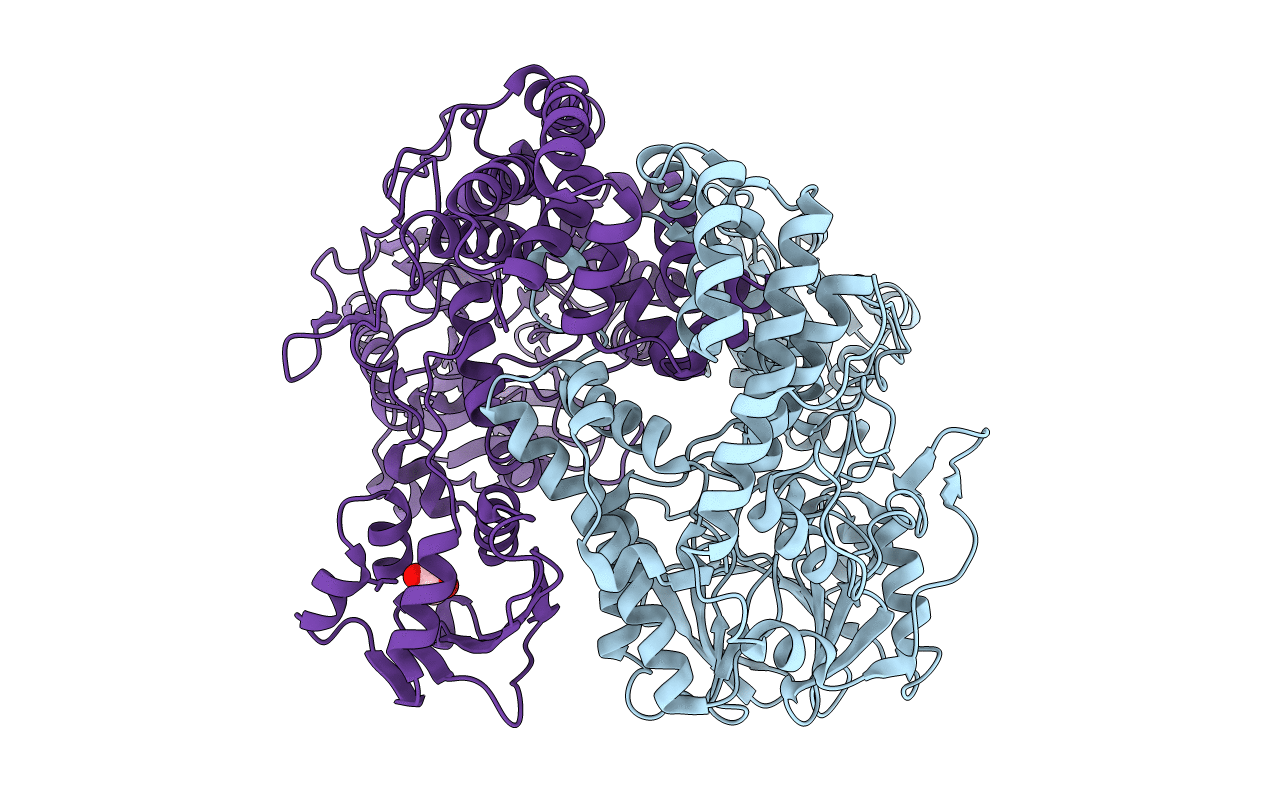
Crystal Structure Of Putataive Short-Chain Dehydrogenase/Reductase (Fabg) From Klebsiella Pneumoniae Subsp. Pneumoniae Ntuh-K2044 In Complex With Nadh
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-03-02 Classification: BIOSYNTHETIC PROTEIN Ligands: K, CL, NAI, GOL, EDO |
 |
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: EDO |
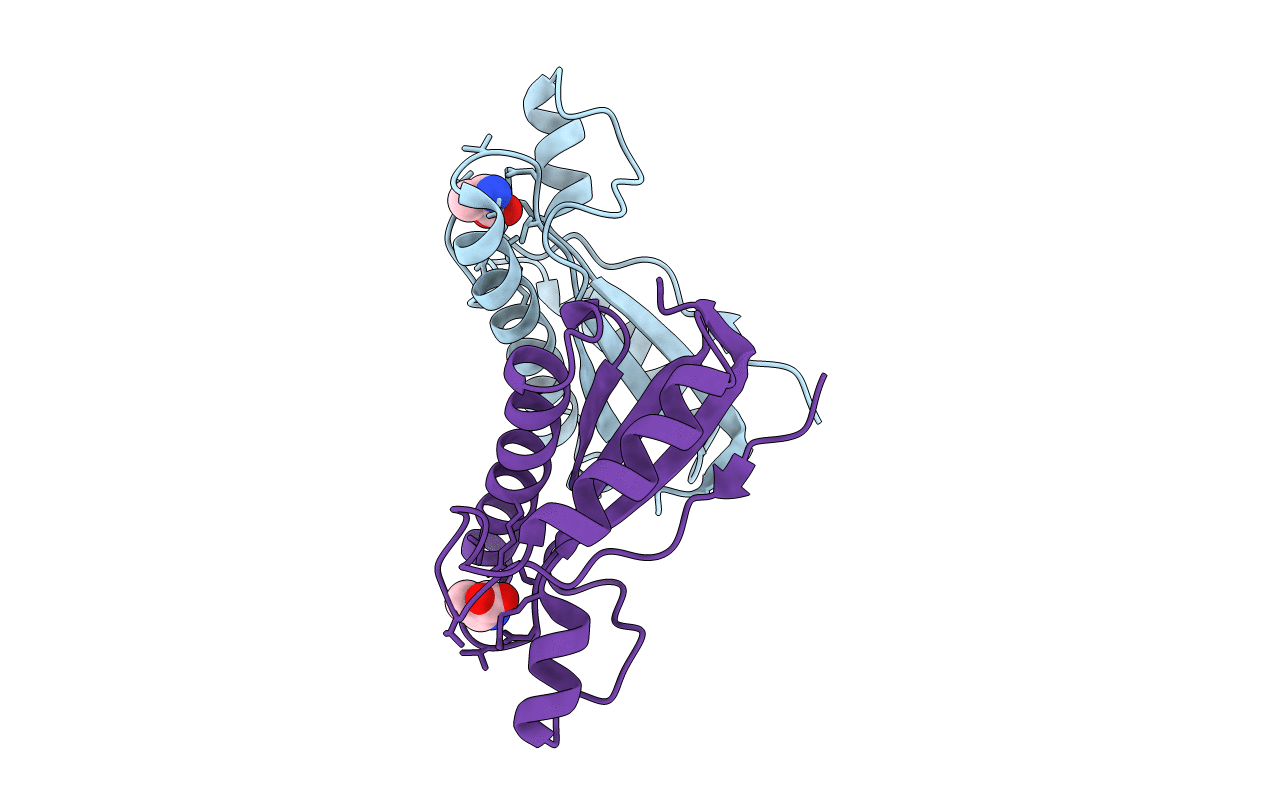 |
Crystal Structure Of The Peptidoglycan Binding Domain Of The Outer Membrane Protein (Ompa) From Klebsiella Pneumoniae With Bound D-Alanine
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-07-28 Classification: PEPTIDE BINDING PROTEIN Ligands: DAL, CL |
 |
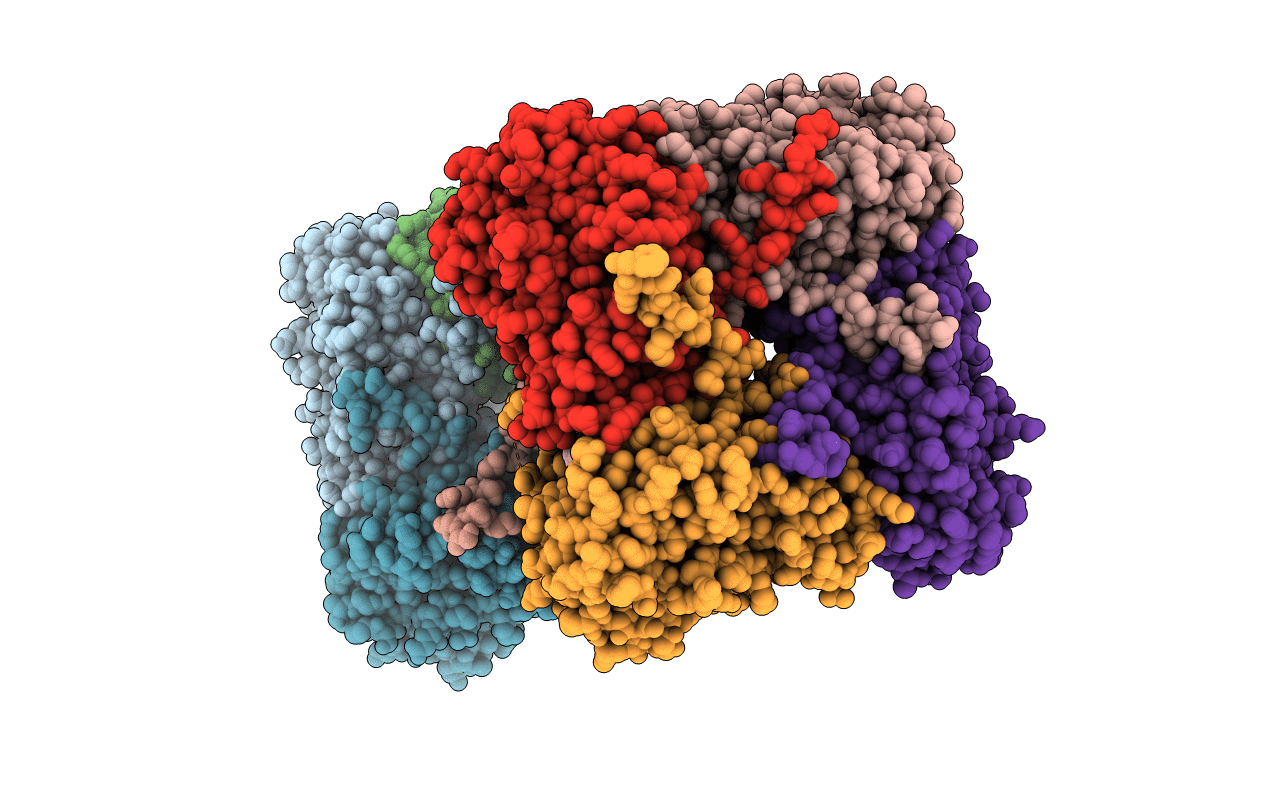
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With Imp And The Inhibitor P221
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, GOL, ZO7, K, EDO |
 |
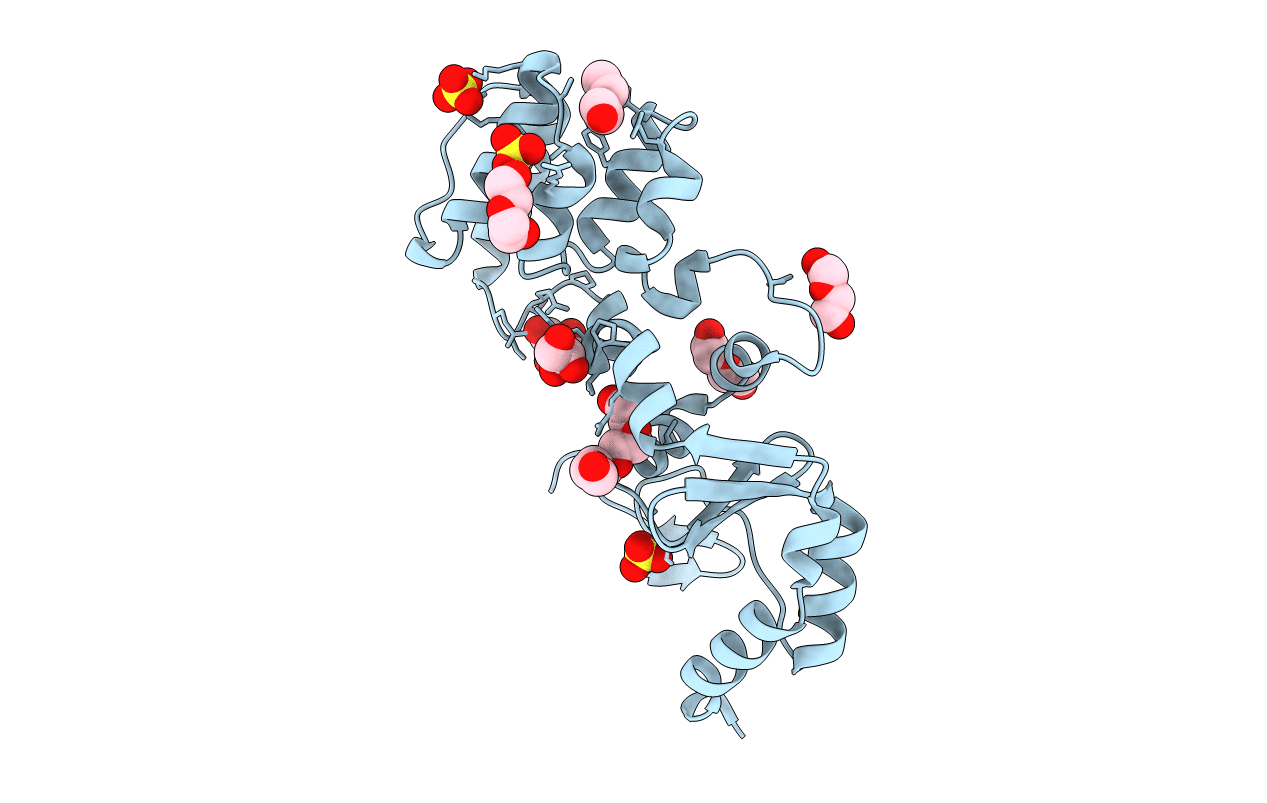
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With Imp And The Inhibitor P176
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: IMP, ZO4, K |
 |
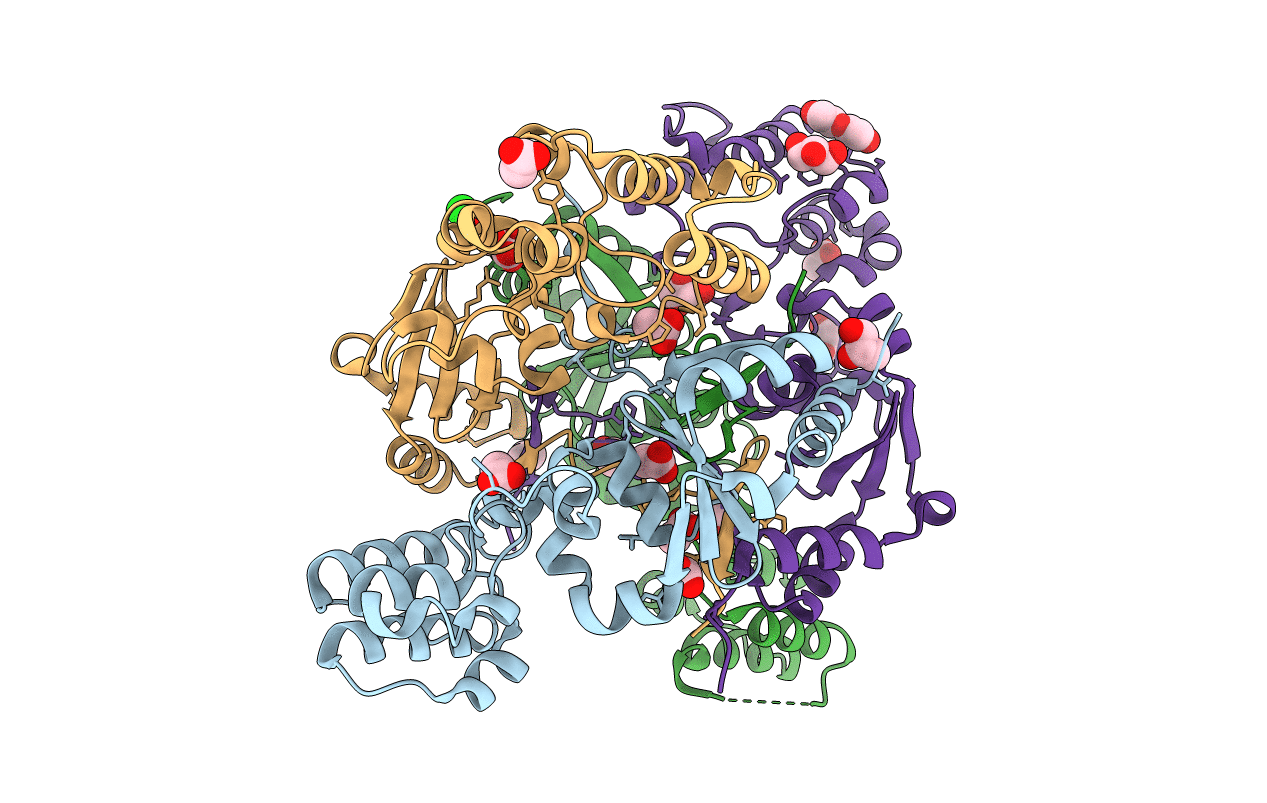
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, N239 Deletion Mutant
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-01-27 Classification: SIGNALING PROTEIN Ligands: GOL, PEG, PG4, SO4 |
 |
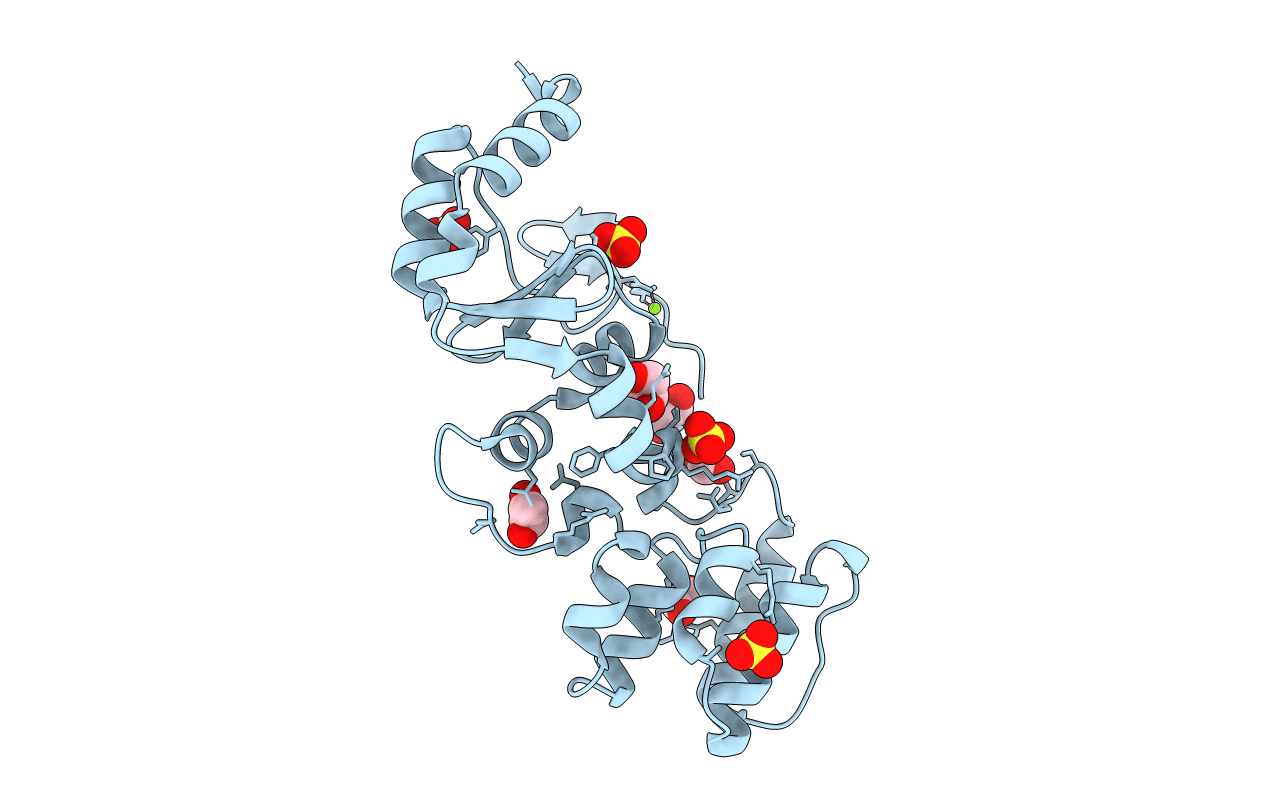
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, 2Nd Form
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-10-14 Classification: SIGNALING PROTEIN Ligands: GOL, ACT, CL, PEG, SRT |
 |
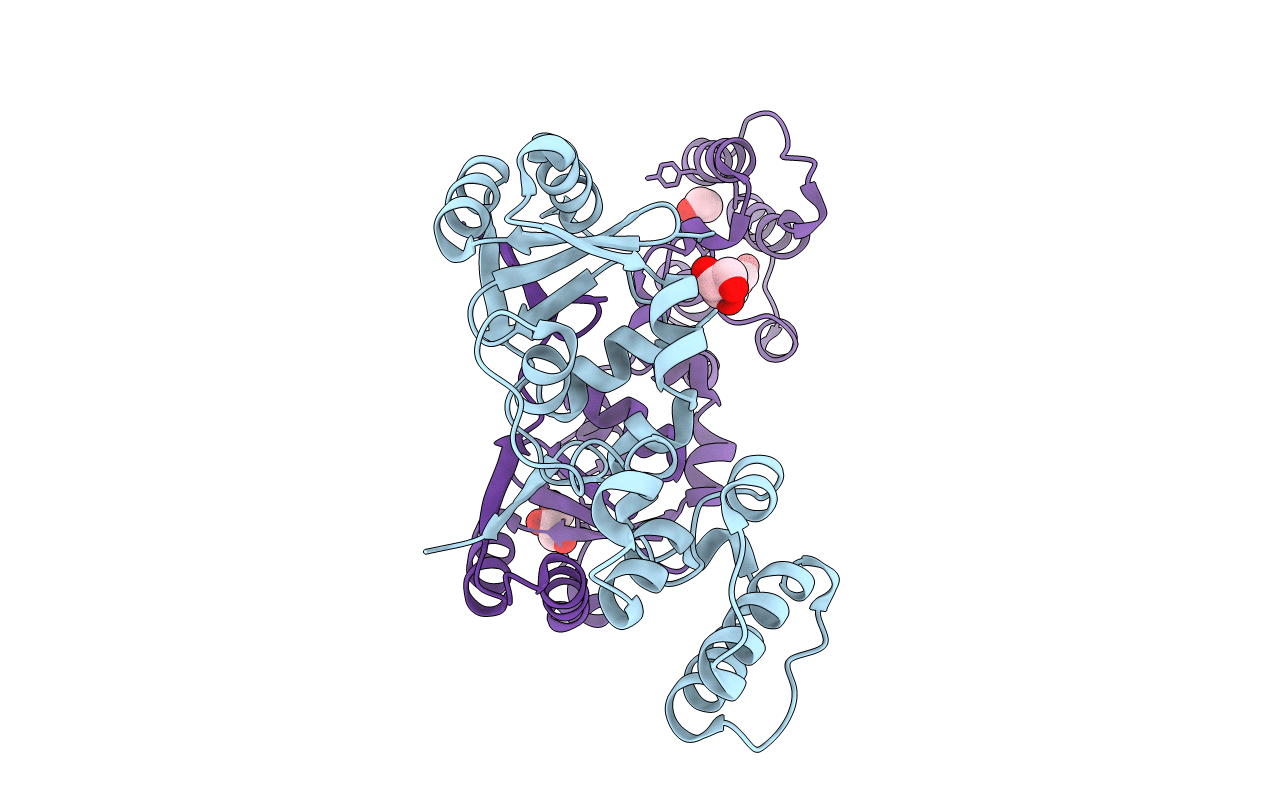
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, N239-T240 Deletion Mutant
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-10-14 Classification: SIGNALING PROTEIN Ligands: EDO, SO4, MG |
 |
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, D238-T240 Deletion Mutant
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-10-14 Classification: SIGNALING PROTEIN Ligands: GOL, EDO |
 |
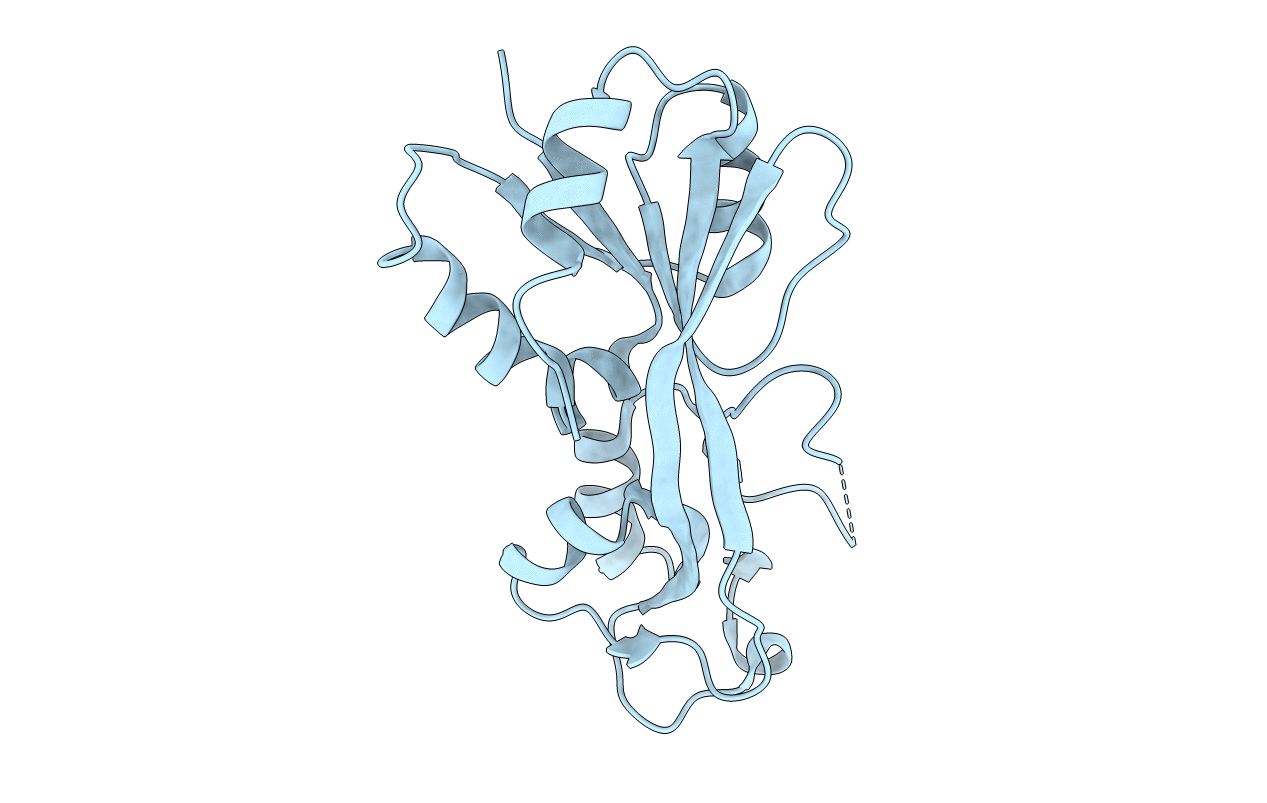
The Crystal Structure Of A Functional Uncharacterized Protein Kp1_0663 From Klebsiella Pneumoniae Subsp. Pneumoniae Ntuh-K2044
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-06-03 Classification: UNKNOWN FUNCTION |
 |
1.52 Angstrom Resolution Crystal Structure Of Transcriptional Regulator Hdfr From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2020-05-06 Classification: TRANSCRIPTION Ligands: CL |
 |
2.70 Angstrom Resolution Crystal Structure Of Uracil Phosphoribosyl Transferase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-05-06 Classification: TRANSFERASE Ligands: BDF, SO4, CL |
 |
Crystal Structure Of The Beta Lactamase Class A Penp From Bacillus Subtilis In The Complex With The Non-Beta- Lactam Beta-Lactamase Inhibitor Avibactam
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Release Date: 2020-03-25 Classification: HYDROLASE Ligands: NXL, FMT, EDO |
 |
The 2.0 A Crystal Structure Of The Type B Chloramphenicol Acetyltransferase From Vibrio Cholerae
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: MPD, EDO, CL, PO4 |
 |
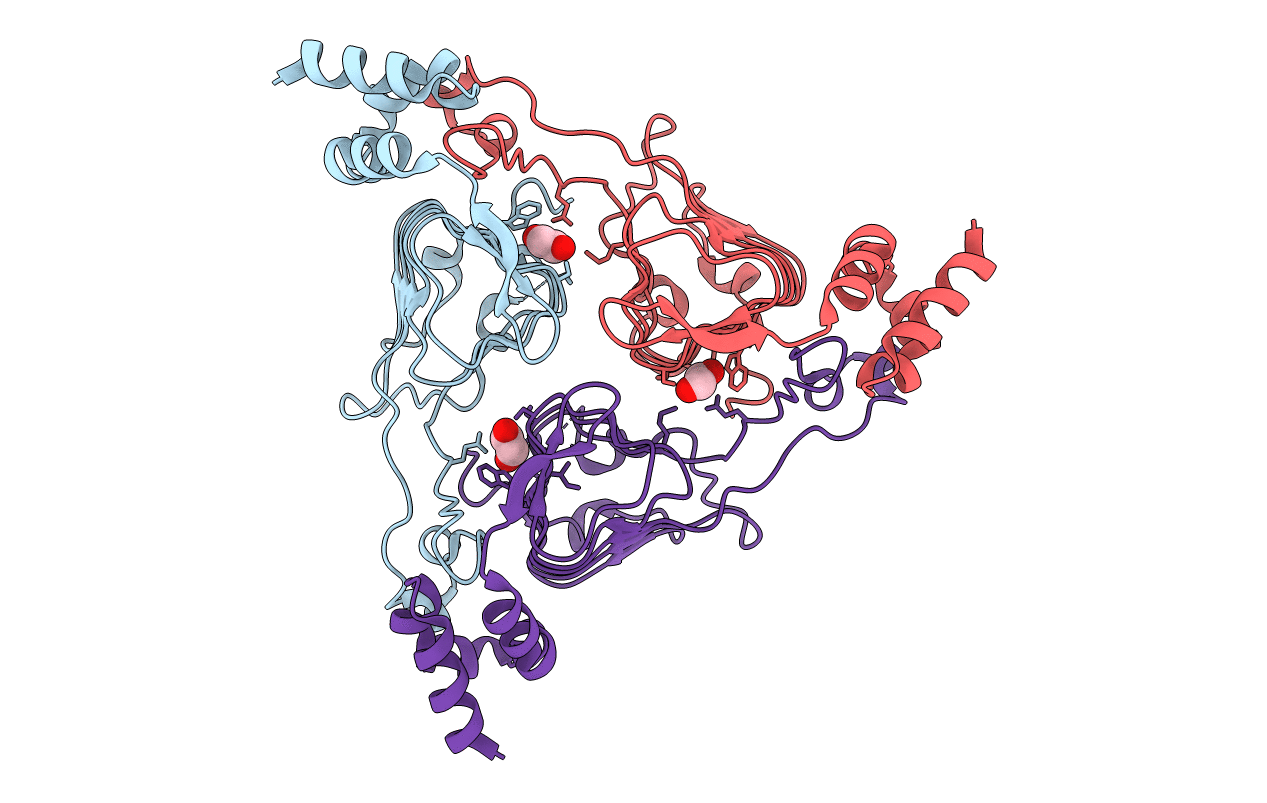
The Crystal Structure Of Chloramphenicol Acetyltransferase-Like Protein From Vibrio Fischeri Es114 In Complex With Taurocholic Acid
Organism: Aliivibrio fischeri (strain atcc 700601 / es114)
Method: X-RAY DIFFRACTION Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: TCH, CL, SO4, ACT, GOL, FMT |
 |
Crystal Structure Of The Type B Chloramphenicol O-Acetyltransferase From Vibrio Vulnificus
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Release Date: 2019-08-14 Classification: TRANSFERASE Ligands: EDO, CL |
 |
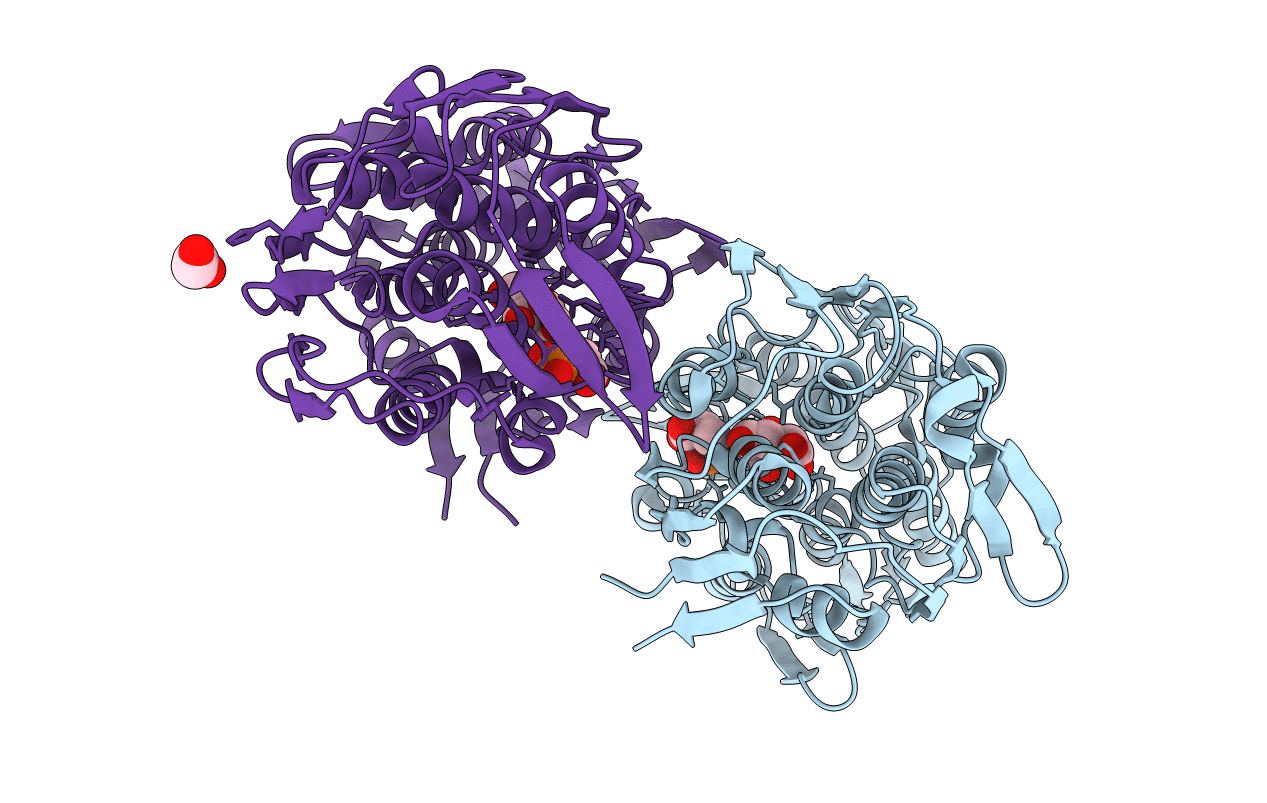
Crystal Structure Of Product-Bound Complex Of Spermidine/Spermine N-Acetyltransferase Speg
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-07-10 Classification: TRANSFERASE Ligands: SPM, SP5, MRD, TRS, MPD, HLG |
 |
1.3 Angstrom Resolution Crystal Structure Of Udp-N-Acetylglucosamine 1-Carboxyvinyltransferase From Streptococcus Pneumoniae In Complex With (2R)-2-(Phosphonooxy)Propanoic Acid.
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: 0V5, CIT, CL, EDO |
 |
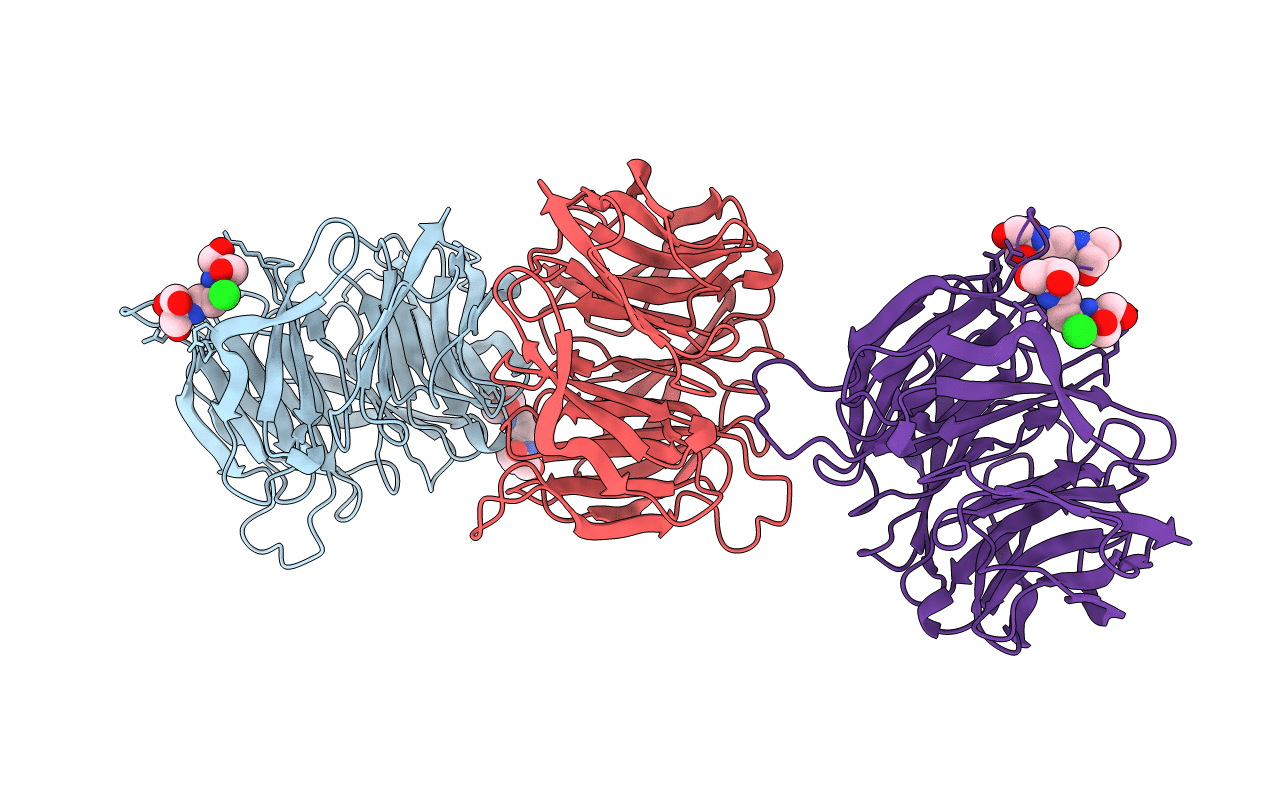
Crystal Structure Of The Sugar Binding Domain Of Laci Family Protein From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-12-26 Classification: SUGAR BINDING PROTEIN |
 |
1.55 Angstrom Resolution Crystal Structure Of 6-Phosphogluconolactonase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-12-19 Classification: HYDROLASE Ligands: B3P, CL |

