Search Count: 115
 |
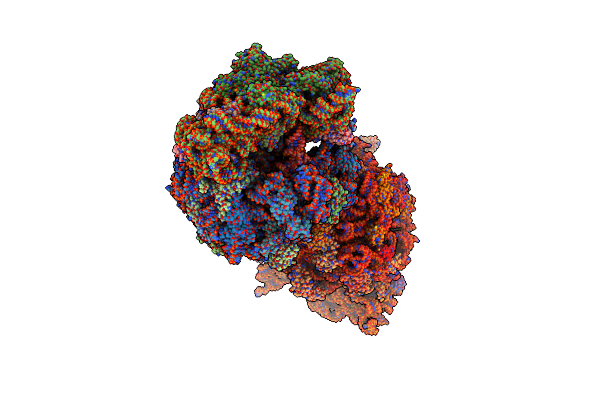
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Spectinomycin, Mrna, Deacylated A- And E-Site Trnaphe, And Deacylated P-Site Trnamet At 2.60A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-08-07 Classification: RIBOSOME/INHIBITOR,ANTIBIOTIC Ligands: MG, K, ZN, SCM, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Spectinomycin Derivative 2694, Mrna, Deacylated A- And E-Site Trnaphe, And Deacylated P-Site Trnamet At 2.75A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-08-07 Classification: RIBOSOME/INHIBITOR Ligands: MG, ZN, Y7K, SF4, K |
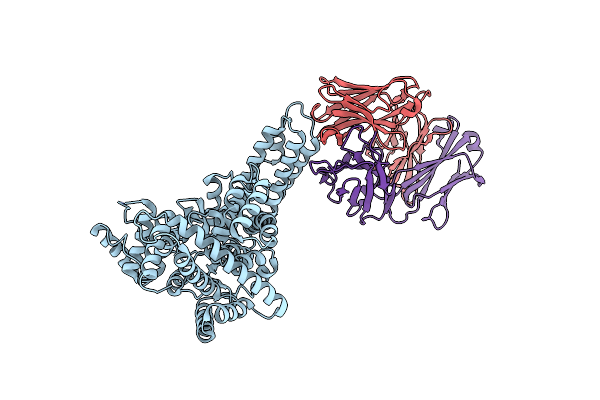 |
Novel Structural Insights For A Pair Of Monoclonal Antibodies Recognizing Non-Overlapping Epitopes Of The Glucosyltransferase Domain Of Clostridium Difficile Toxin B
Organism: Homo sapiens, Clostridioides difficile r20291
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-05-11 Classification: TOXIN/IMMUNE SYSTEM |
 |
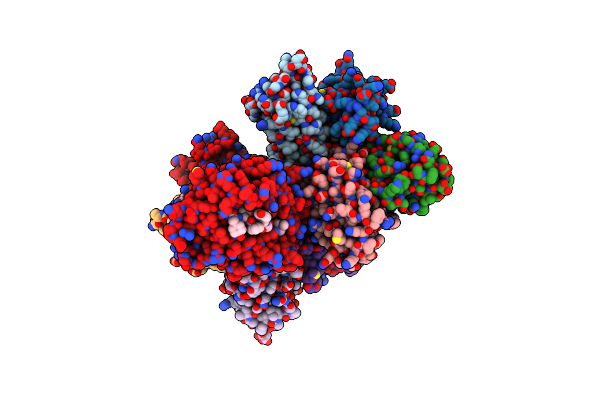
Novel Structural Insights For A Pair Of Monoclonal Antibodies Recognizing Non-Overlapping Epitopes Of The Glucosyltransferase Domain Of Clostridium Difficile Toxin B
Organism: Clostridioides difficile, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2022-05-11 Classification: TOXIN/IMMUNE SYSTEM |
 |
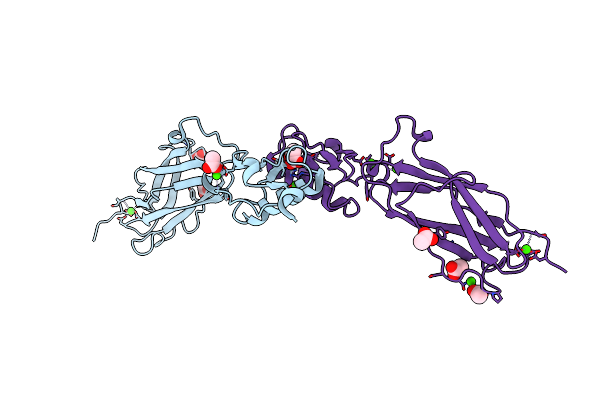
Noncanonical Binding Of Single-Chain A6 Tcr Variant S3-4 In Complex With Tax/Hla-A2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2020-10-28 Classification: IMMUNE SYSTEM Ligands: MES |
 |
Organism: Streptococcus sanguinis (strain sk36)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-01-27 Classification: SUGAR BINDING PROTEIN Ligands: CA, ACT |
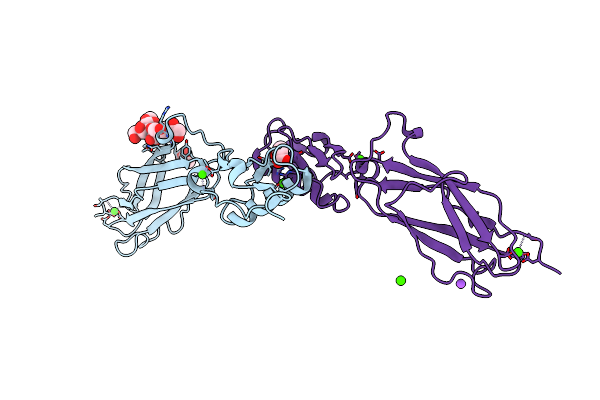 |
Crystal Structure Of The Srpa Adhesin From Streptococcus Sanguinis With A Sialyl Galactose Disaccharide Bound
Organism: Streptococcus sanguinis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-27 Classification: SUGAR BINDING PROTEIN Ligands: CA, ACT, NA |
 |
Crystal Structure Of The Srpa Adhesin R347E Mutant From Streptococcus Sanguinis
Organism: Streptococcus sanguinis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-01-27 Classification: SUGAR BINDING PROTEIN Ligands: CA, ACT |
 |
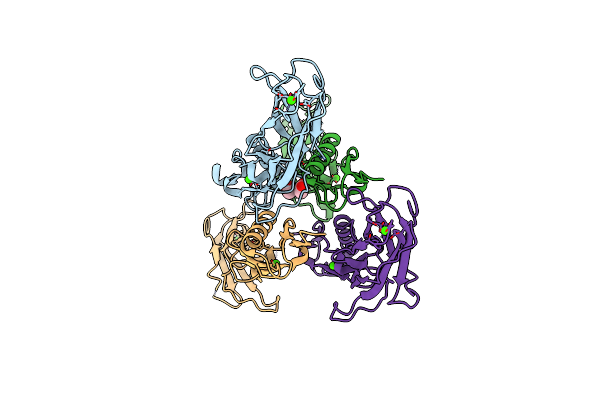
Ternary Complex Of Y-Family Dna Polymerase Dpo4 With (5'S)-8,5'-Cyclo-2'-Deoxyguanosine And Dttp
Organism: Sulfolobus solfataricus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2015-01-14 Classification: TRANSFERASE/DNA Ligands: MG, DOC, CA, TTP |
 |
Ternary Complex Of Y-Family Dna Polymerase Dpo4 With (5'S)-8,5'-Cyclo-2'-Deoxyguanosine And Dctp
Organism: Sulfolobus solfataricus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2015-01-14 Classification: TRANSFERASE/DNA Ligands: DCP, MG, DOC |
 |
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: CA, SO4 |
 |
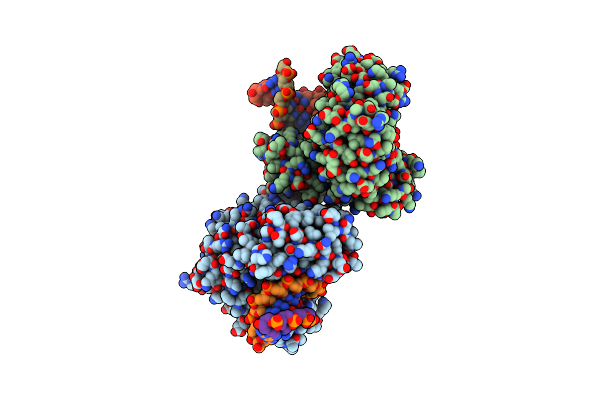
Organism: Homo sapiens, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-05-28 Classification: PROTEIN BINDING Ligands: NAG |
 |
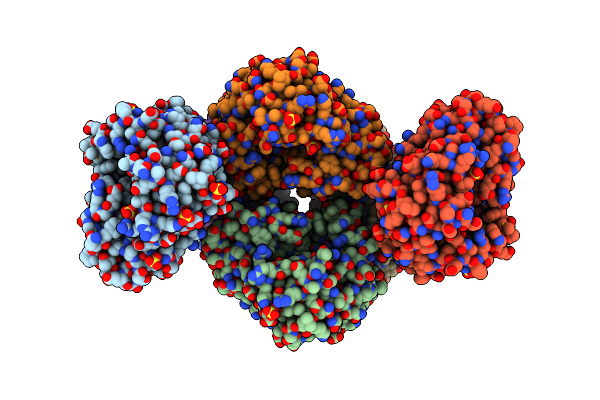
2.60 Angstrom Resolution Crystal Structure Of A Protein Kinase Domain Of Type Iii Effector Nleh2 (Ecs1814) From Escherichia Coli O157:H7 Str. Sakai
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: GOL, PEG |
 |
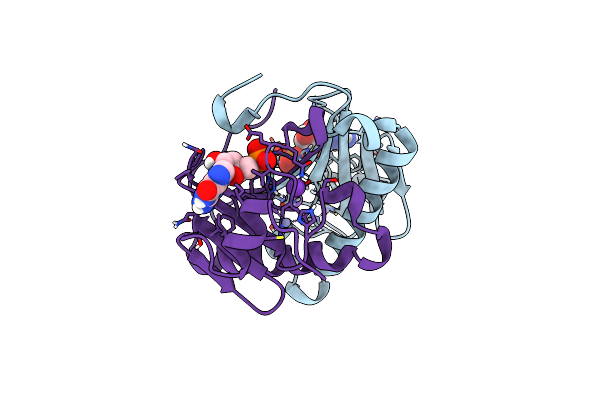
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
0.95A Resolution Structure Of A Histidine Triad Protein From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2012-04-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: 5GP, ZN, K |
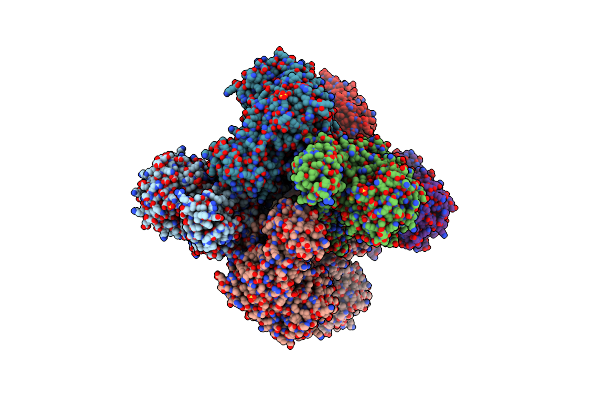 |
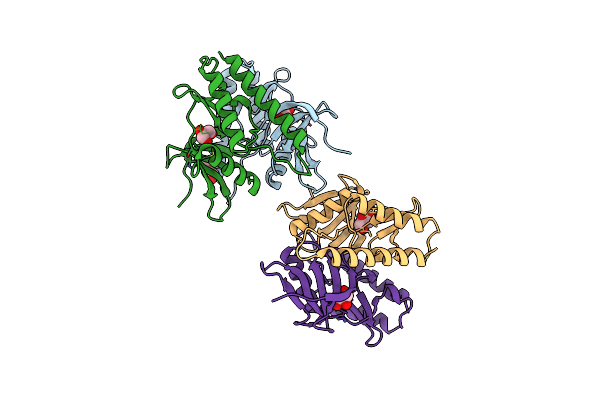
The Crystal Structure Of Sucrose Synthase-1 From Arabidopsis Thaliana And Its Functional Implications.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2011-08-24 Classification: TRANSFERASE Ligands: UDP, FRU, SO4, MLA, K |
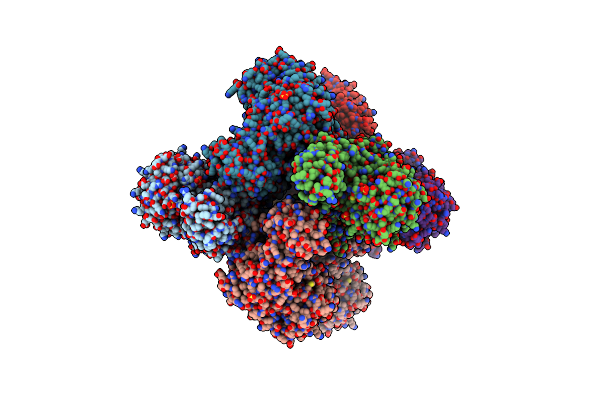 |
The Crystal Structure Of Sucrose Synthase-1 In Complex With A Breakdown Product Of The Udp-Glucose
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-08-24 Classification: TRANSFERASE Ligands: UDP, LCN, NHF, SO4, MLA, K |
 |
The Crystal Structure Of Sucrose Synthase-1 From Arabidopsis Thaliana And Its Functional Implications.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2011-08-24 Classification: TRANSFERASE Ligands: UDP, FRU, SO4, MLA, K |
 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-10 Classification: LYASE |
 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-07-20 Classification: TRANSFERASE Ligands: SO4, K |

