Search Count: 99
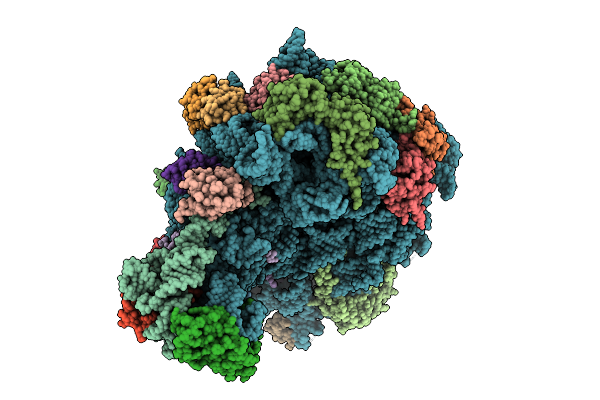 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
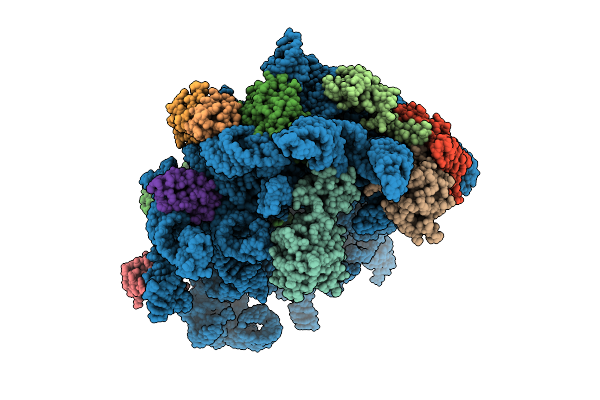 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
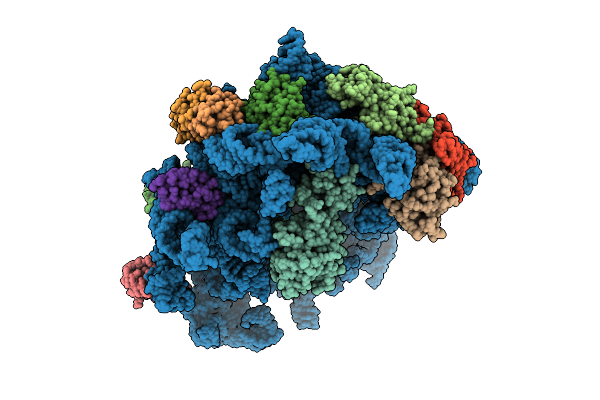 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: AN6 |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: WHC, GOL, ZN |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: WHC, GOL, ZN |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: GOL, ZN, ACT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, ATP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: D06 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: D06, TMO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: CW8 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: CW8, TMO |
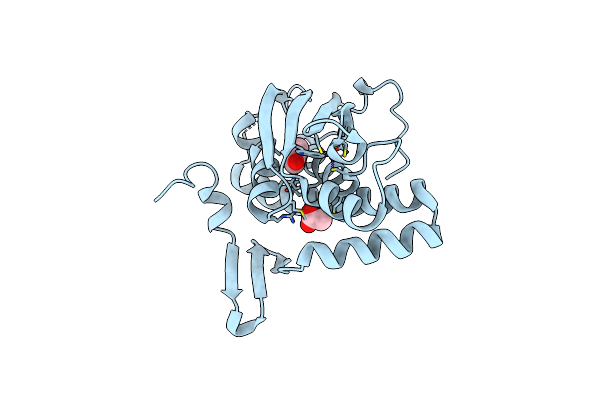 |
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-09-07 Classification: DNA BINDING PROTEIN Ligands: IPH, ACT, ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-01-19 Classification: LIGASE Ligands: ADP, GOL, SO4, MG |

