Search Count: 86
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: HYDROLASE Ligands: A1B1V, ZN, CA, NHE, EDO, PEG, PGE, PG4 |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: A1D92 |
 |
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:3.68 Å Release Date: 2025-05-07 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |
 |
Cryoem Structure Of Bchn-Bchb Bound To Pchlide From The Dpor Under Turnover Complex Dataset
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:3.29 Å Release Date: 2025-05-07 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |
 |
Cryoem Structure Of Bchn-Bchb Electron Acceptor Component Protein Of Dpor With Pchlide
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:2.92 Å Release Date: 2025-05-07 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |
 |
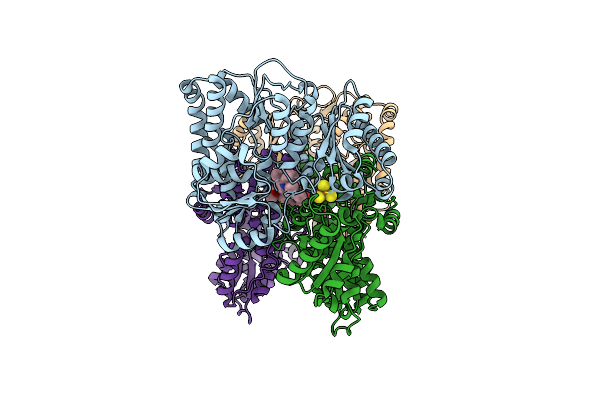
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: SF4, CU |
 |
Cryoem Structure Of Bchn-Bchb Electron Acceptor Component Protein Of Dpor With Pchlide
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:3.13 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |
 |
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU, MG |
 |
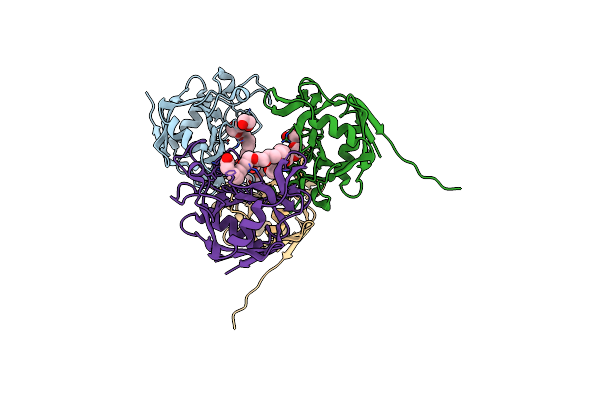 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase Containing (2S,3S)-4-Fluorovaline
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-10-09 Classification: ISOMERASE Ligands: 1PE, EDO, PGE, ACT, PEG, XPE |
 |
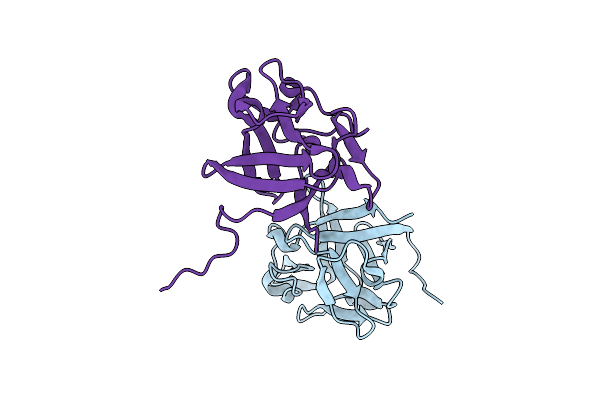
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-06-26 Classification: SIGNALING PROTEIN |
 |
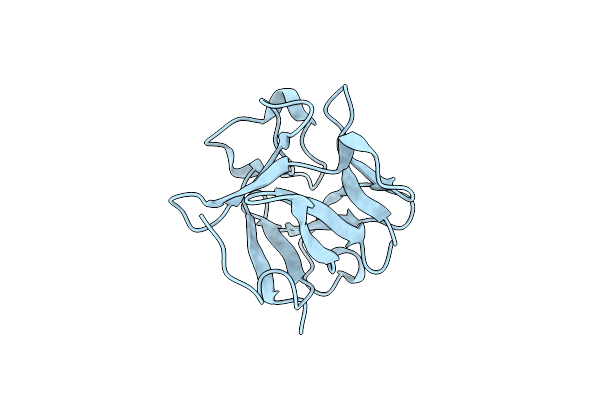
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-06-26 Classification: SIGNALING PROTEIN |
 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase Containing Delta1-Monofluoro-Leucines
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-05-22 Classification: ISOMERASE |
 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase Containing Delta2-Monofluoro-Leucines
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-05-22 Classification: ISOMERASE Ligands: EDO, PGE, PEG, 1PE, XPE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-05-22 Classification: ISOMERASE Ligands: PEG, PGE, EDO, PG4, 2PE |
 |
Organism: Acinetobacter
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-19 Classification: HYDROLASE |
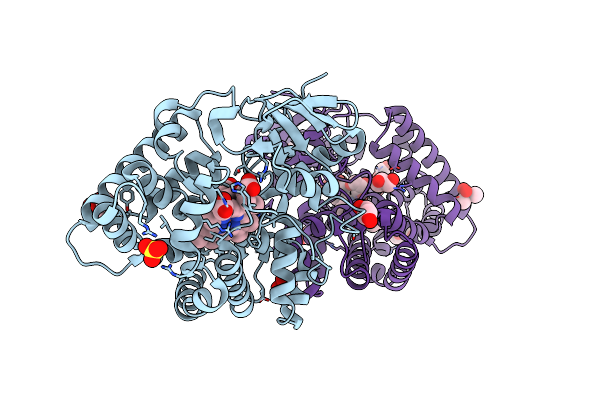 |
Crystal Structure Of Cytochrome P450 Pikc With The Unnatural Amino Acid P-Acetyl-L-Phenylalanine Incorporated At Position 238
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, CAC, GOL, PEG |
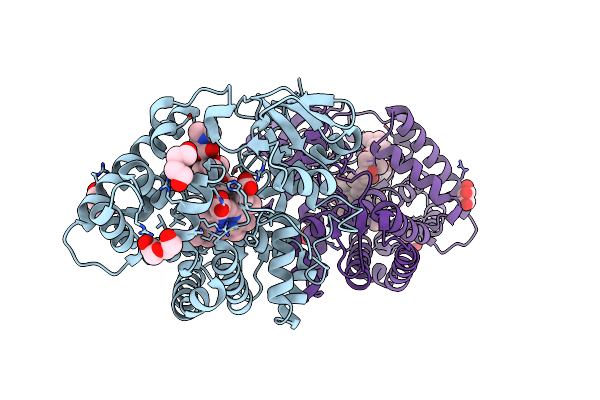 |
Crystal Structure Of Yc-17-Bound Cytochrome P450 Pikc With The Unnatural Amino Acid P-Acetyl-L-Phenylalanine Incorporated At Position 238
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: HEM, PXI, GOL, PEG |
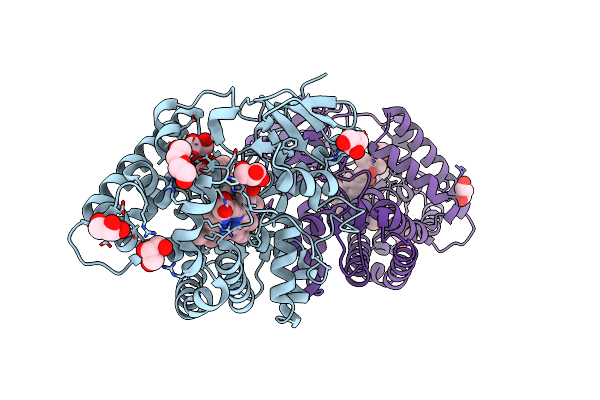 |
Crystal Structure Of 10-Dml-Bound Cytochrome P450 Pikc With The Unnatural Amino Acid P-Acetyl-L-Phenylalanine Incorporated At Position 238
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: HEM, E4H, GOL, PEG |
 |
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: 0TL, CA |

