Search Count: 1,680
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: PROTEIN BINDING |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG, DD9, D10, HP6, XKP, PEF, PLM, DCR |
 |
Organism: Novosphingobium sp. 28-62-57
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GDP |
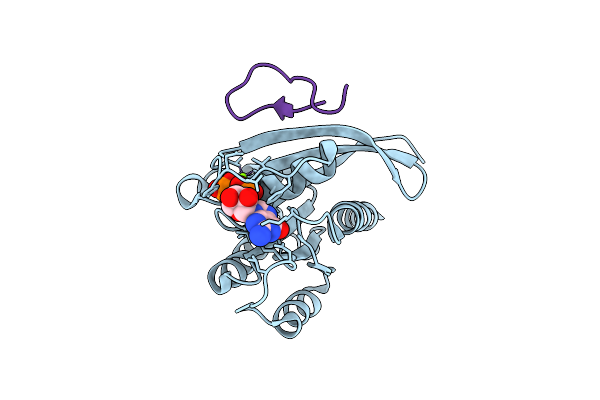 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GTP |
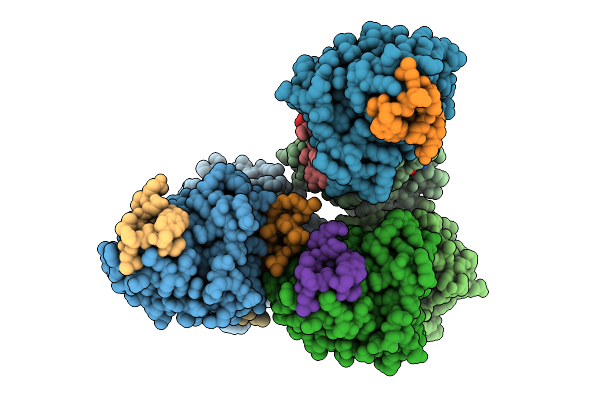 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GTP |
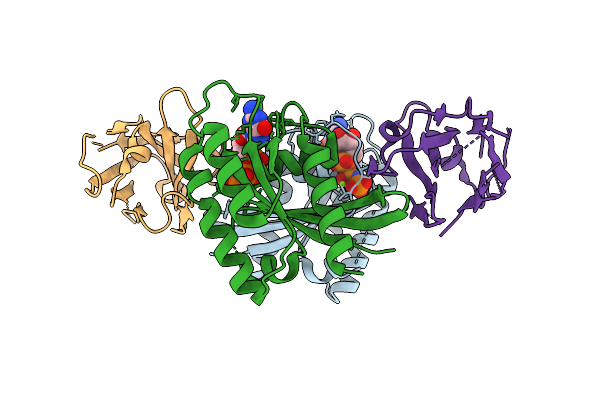 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GNP |
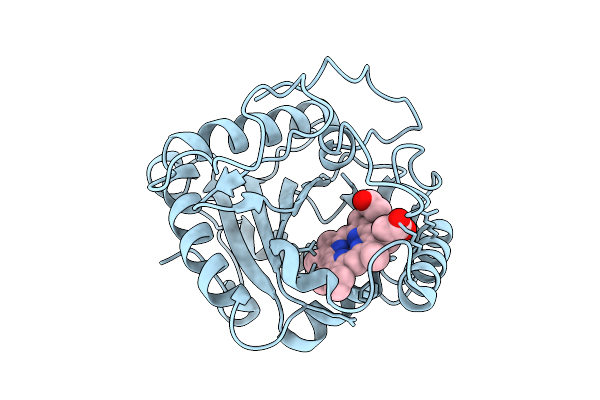 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: ARG, HEM |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG, PGL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: 8K3 |
 |
Organism: Ectopseudomonas mendocina ymp
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: TRS, GOL, CA, EDO |
 |
Organism: [haemophilus] ducreyi
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSFERASE Ligands: NMN |
 |
Organism: [haemophilus] ducreyi
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSFERASE Ligands: NCA |
 |
Organism: Rattus norvegicus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CELL CYCLE Ligands: GTP, MG, CA, MES, W4Q, GDP, ACP |

