Search Count: 3,255
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: CO, A1CZS |
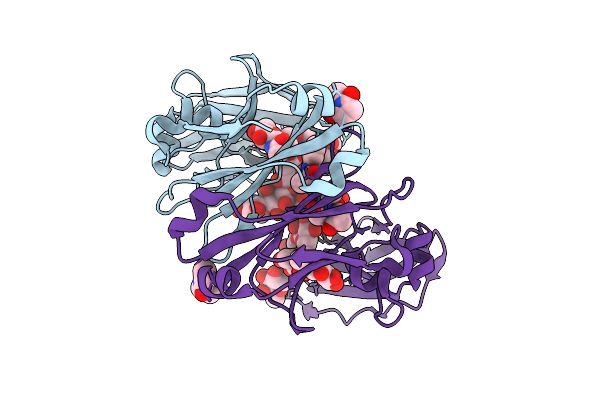 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: A1CZS |
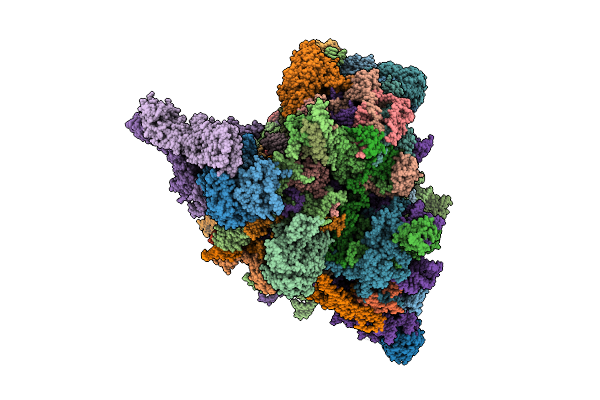 |
Translational Activators Aep1, Aep2 And Atp25 In Complex With Mrna And The Yeast Mitochondrial Ribosome
Organism: Saccharomyces cerevisiae w303
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: MG, GTP |
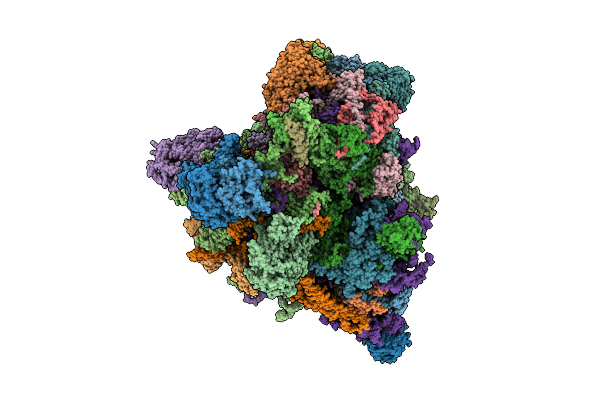 |
Translational Activator Aep3 In Complex With Mrna And The Yeast Mitochondrial Ribosome
Organism: Saccharomyces cerevisiae w303, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: MG, GTP |
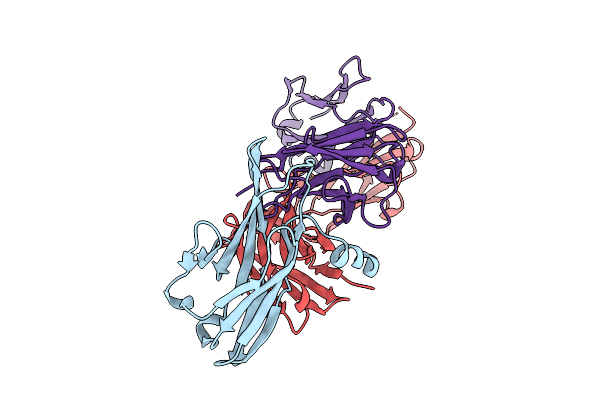 |
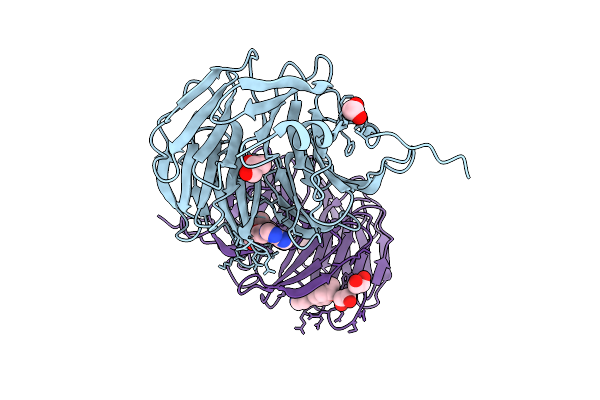
Crystal Structure Of Pilra In Complex With Fab Portion Of Antagonist Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: NUCLEAR PROTEIN Ligands: GOL, A1BI5, EDO |
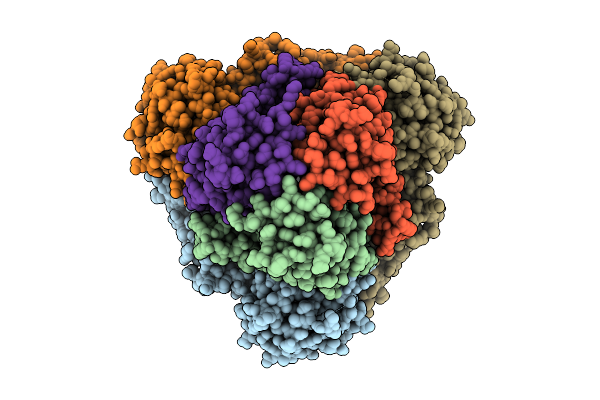 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: FE2, FES, HCI |
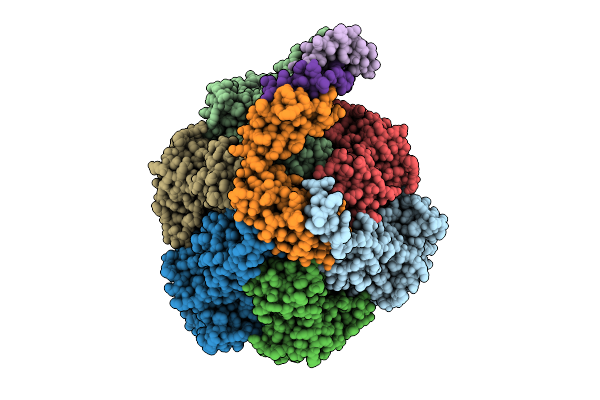 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, PO4 |
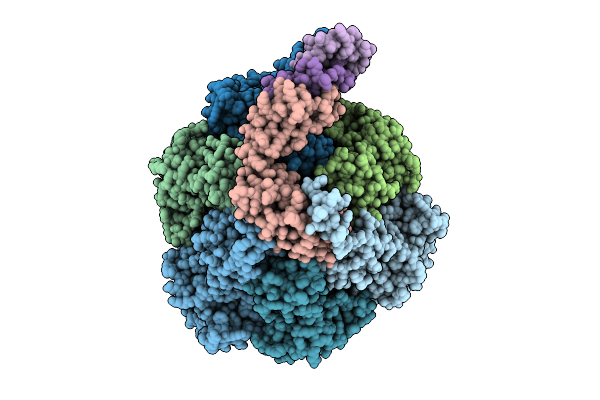 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: UNKNOWN FUNCTION |

