Search Count: 182
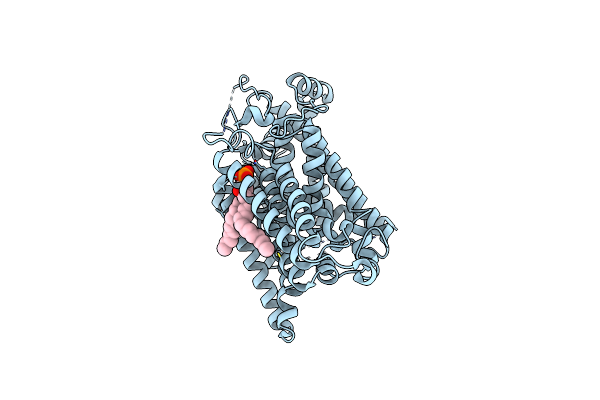 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: TRANSPORT PROTEIN Ligands: 3PH |
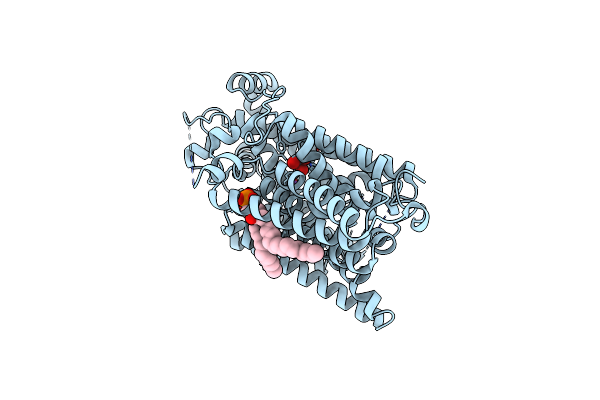 |
Structure Of The Sodium/Iodide Symporter (Nis) In Complex With Perrhenate And Sodium
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: TRANSPORT PROTEIN |
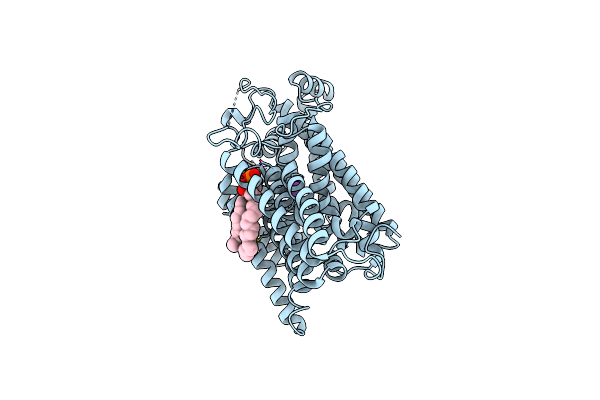 |
Structure Of The Sodium/Iodide Symporter (Nis) In Complex With Iodide And Sodium
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: TRANSPORT PROTEIN |
 |
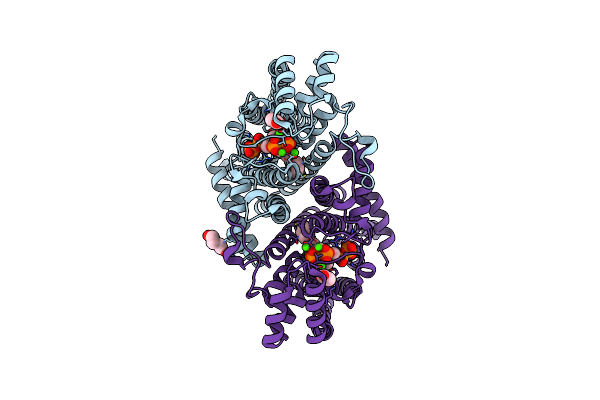
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-05-04 Classification: IMMUNE SYSTEM Ligands: SO4, PG0, NA, CL |
 |
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: TRANSFERASE Ligands: CA, IPR, 476, ACT, PEG |
 |
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-10-07 Classification: TRANSFERASE Ligands: CA, ACT, IPR, 476, PEG |
 |
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-10-07 Classification: TRANSFERASE Ligands: CA, IPR, 476, ACT |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2019-09-11 Classification: CYTOSOLIC PROTEIN |
 |
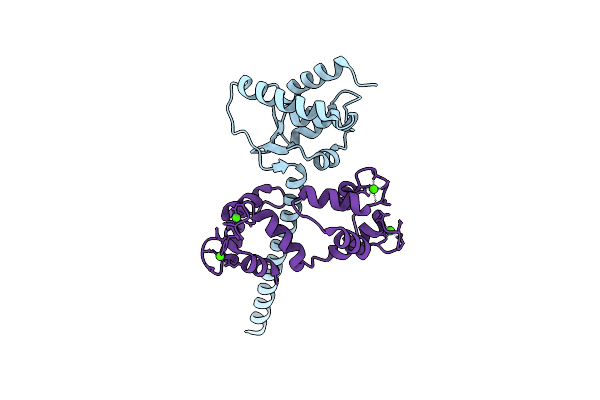
Crystal Structure Of Human Nav1.4 Cterminal Domain In Complex With Apo Calmodulin
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-10 Classification: CALMODULIN-BINDING PROTEIN Ligands: CL, EDO, CO3, TRS |
 |
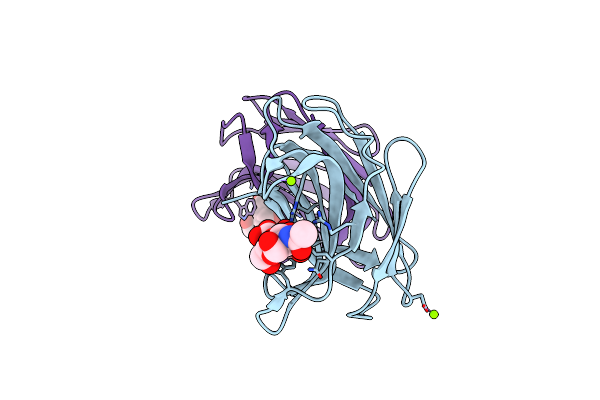
Crystal Structure Of Human Nav1.4 C-Terminal (1599-1754) Domain In Complex With Calcium-Bound Calmodulin
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-04-10 Classification: PROTEIN BINDING Ligands: CA |
 |
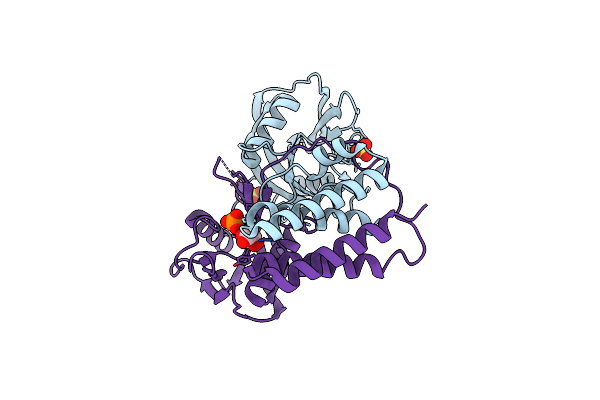
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-03-20 Classification: SUGAR BINDING PROTEIN Ligands: MG |
 |
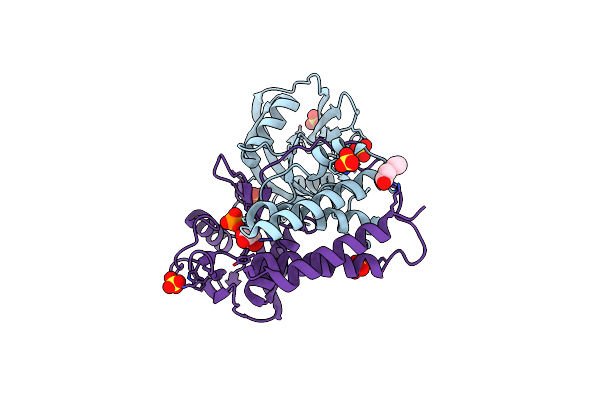
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: SO4, PO4, RB5 |
 |
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: SO4, PO4, PEG, RB5 |
 |
Crystal Structure Of H108A Peptidylglycine Alpha-Hydroxylating Monooxygenase (Phm)
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: CU, NI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2019-02-06 Classification: TRANSFERASE Ligands: 144, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2019-01-30 Classification: TRANSFERASE Ligands: MN, PGE, GOL, VO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-08-22 Classification: TRANSFERASE Ligands: SO4, MN |
 |
Crystal Structure Of H107A Peptidylglycine Alpha-Hydroxylating Monooxygenase (Phm) In Complex With Citrate
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-07-18 Classification: OXIDOREDUCTASE Ligands: CU, GOL, NI, FLC |
 |
Crystal Structure Of Apo Wild Type Peptidylglycine Alpha-Hydroxylating Monooxygenase (Phm)
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-07-18 Classification: OXIDOREDUCTASE Ligands: PEG, GOL |
 |
Crystal Structure Of Apo Wild Type Peptidylglycine Alpha-Hydroxylating Monooxygenase (Phm) Soaked With Peptide (Peptide Not Observed)
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-07-18 Classification: OXIDOREDUCTASE |

