Search Count: 18
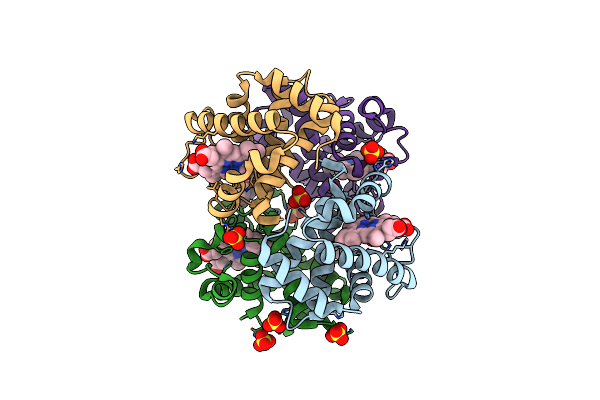 |
Crystal Structure Of Ferric R-State Human Methemoglobin Bound To Maleimide-Deferoxamine Bifunctional Chelator (Dfo)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-09-18 Classification: OXYGEN TRANSPORT Ligands: HEM, OXY, SO4, G7Z |
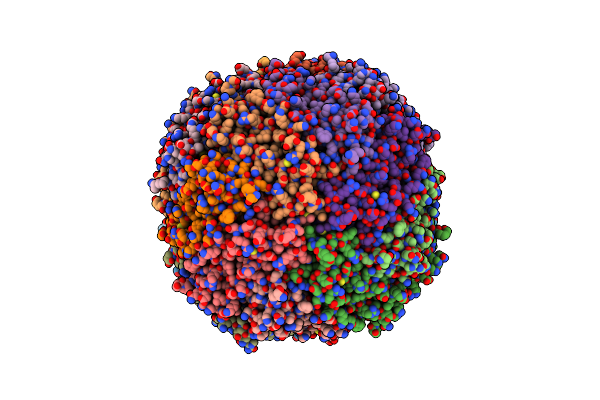 |
Structure Of A Modified Mouse H-Chain Ferritin With A Lanthanide Binding Motif
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2018-07-25 Classification: METAL TRANSPORT Ligands: FE |
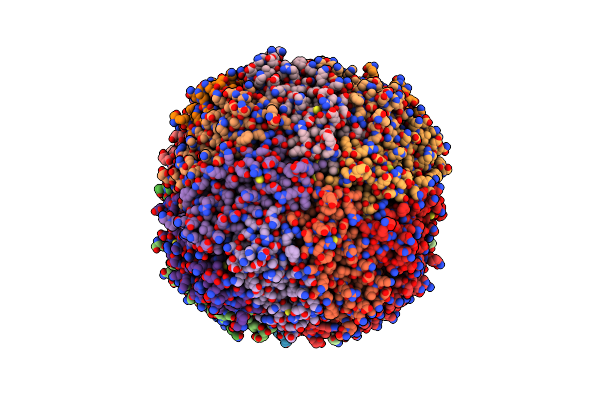 |
Structure Of A Modified Mouse H Chain Ferritin With A Lanthanide Binding Motif In Complex With Terbium
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2018-07-25 Classification: METAL TRANSPORT Ligands: TB |
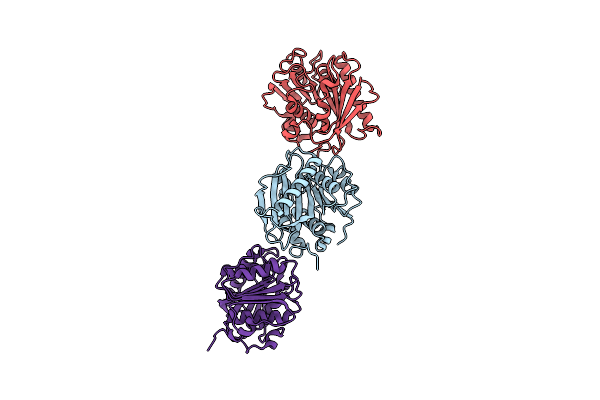 |
Crystal Structure Of A Polyethylene Terephthalate Degrading Hydrolase From Ideonella Sakaiensis Collected At Long Wavelength
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CL |
 |
High Resolution Crystal Structure Of A Polyethylene Terephthalate Degrading Hydrolase From Ideonella Sakaiensis
Organism: Ideonella sakaiensis (strain 201-f6)
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CL, NA |
 |
Crystal Structure Of A Polyethylene Terephthalate Degrading Hydrolase From Ideonella Sakaiensis In Spacegroup P212121
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of A Polyethylene Terephthalate Degrading Hydrolase From Ideonella Sakaiensis In Spacegroup P21
Organism: Ideonella sakaiensis (strain 201-f6)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CL, SO4 |
 |
Crystal Structure Of A Polyethylene Terephthalate Degrading Hydrolase From Ideonella Sakaiensis In Spacegroup C2221
Organism: Ideonella sakaiensis (strain 201-f6)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2018-04-25 Classification: HYDROLASE |
 |
Organism: Hypocrea atroviridis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: NAG, NI, GOL, BTB, GS1, GLC, PEG |
 |
Organism: Hypocrea atroviridis (strain atcc 20476 / imi 206040)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: NAG, NI, BTB, GOL, CL, PEG |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2016-11-30 Classification: METAL BINDING PROTEIN Ligands: MG |
 |
Organism: Penicillium funiculosum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-22 Classification: HYDROLASE Ligands: NAG, K, NA, CL |
 |
Organism: Thermobifida fusca tm51
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2014-09-17 Classification: OXIDOREDUCTASE Ligands: HEM, ACT |
 |
Organism: Thalictrum flavum
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2008-08-05 Classification: LYASE |
 |
X-Ray Structure Of Norcoclaurine Synthase From Thalictrum Flavum In Complex With Dopamine And Hydroxybenzaldehyde
Organism: Thalictrum flavum
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2008-08-05 Classification: LYASE Ligands: ACT, CL, HBA, LDP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2008-01-22 Classification: OXIDOREDUCTASE Ligands: FAD, NA |
 |
X-Ray Structure Of A Novel Thermostable Hemoglobin From The Actinobacterium Thermobifida Fusca
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2005-07-20 Classification: OXYGEN STORAGE/TRANSPORT Ligands: HEM, ACT |
 |
The X-Ray Structure Of Ferric Escherichia Coli Flavohemoglobin Reveals An Unespected Geometry Of The Distal Heme Pocket
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2002-08-06 Classification: OXIDOREDUCTASE Ligands: FAD, HEM, NA, CL |

