Search Count: 36
 |
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-02-07 Classification: PLANT PROTEIN Ligands: ACT, SO4 |
 |
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2024-02-07 Classification: PLANT PROTEIN Ligands: ACT, GOL, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-10-04 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Bacillus thuringiensis serovar andalousiensis bgsc 4aw1
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2022-04-13 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, U2F |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-04-13 Classification: TRANSFERASE Ligands: UPG |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-04-13 Classification: BIOSYNTHETIC PROTEIN Ligands: UPG, MG |
 |
Crystal Structure Of Wild-Type Kras4B(1-169) In Complex With Gmppnp And Mg Ion
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-02-10 Classification: ONCOPROTEIN Ligands: GNP, MG |
 |
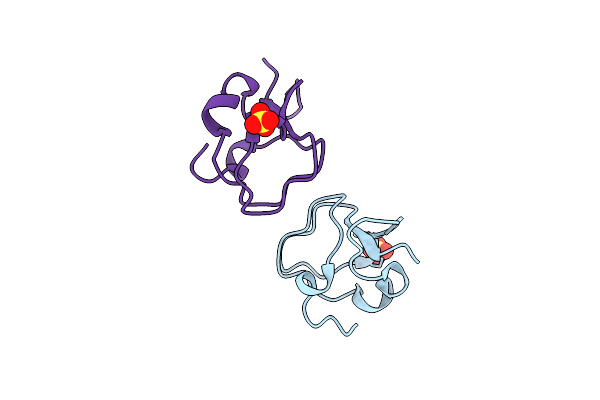
Organism: Staphylococcus warneri
Method: SOLUTION NMR Release Date: 2018-11-28 Classification: ANTIBIOTIC |
 |
Organism: Staphylococcus warneri
Method: SOLUTION NMR Release Date: 2018-11-28 Classification: ANTIBIOTIC |
 |
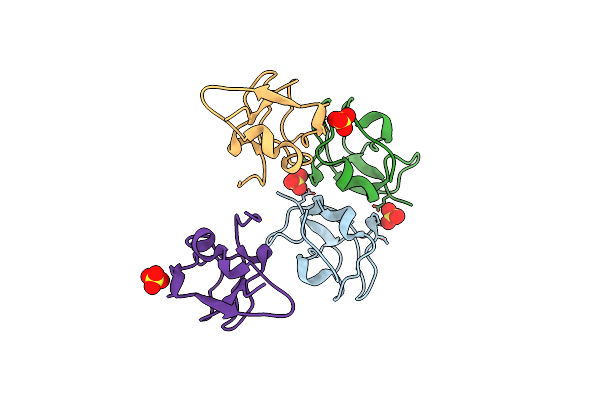
Crystal Structure Of Notched-Fin Eelpout Type Iii Antifreeze Protein (Nfe6, Afp), C2221 Form.
Organism: Zoarces elongatus
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2018-05-16 Classification: ANTIFREEZE PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Notched-Fin Eelpout Type Iii Antifreeze Protein (Nfe6, Afp), P212121 Form
Organism: Zoarces elongatus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2018-05-16 Classification: ANTIFREEZE PROTEIN Ligands: SO4 |
 |
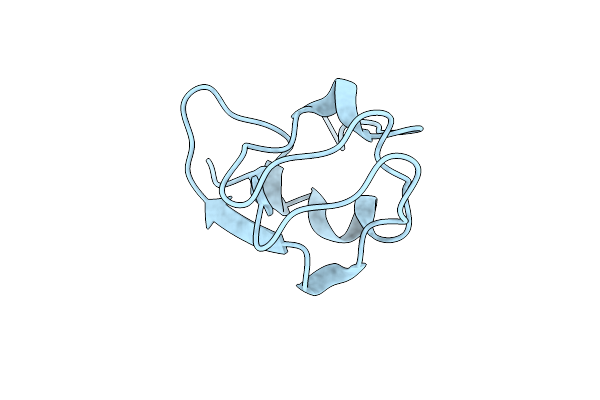
Crystal Structure Of Notched-Fin Eelpout Type Iii Antifreeze Protein A20V Mutant (Nfe6, Afp), C2221 Form
Organism: Zoarces elongatus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-05-16 Classification: ANTIFREEZE PROTEIN Ligands: ACT |
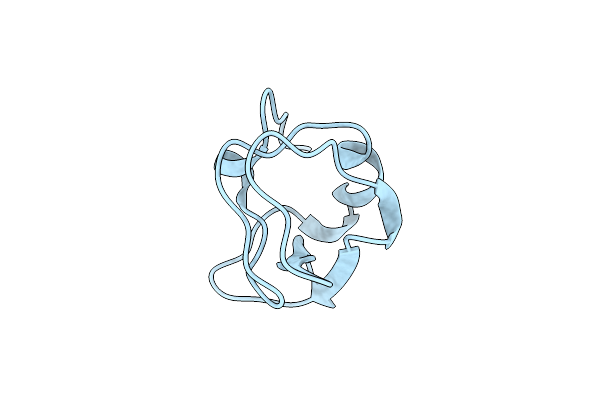 |
Crystal Structure Of Notched-Fin Eelpout Type Iii Antifreeze Protein A20I Mutant (Nfe6, Afp), P212121 Form
Organism: Zoarces elongatus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2018-05-16 Classification: ANTIFREEZE PROTEIN |
 |
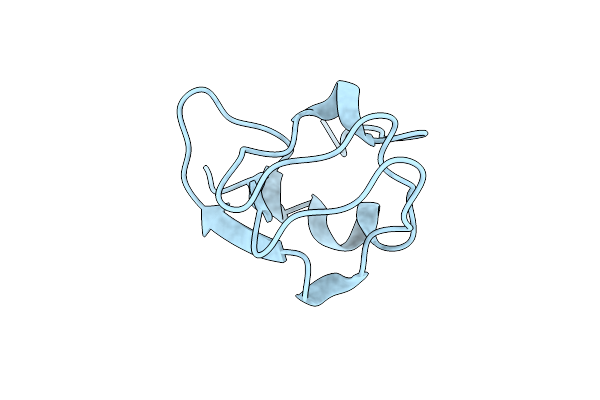
Crystal Structure Of Notched-Fin Eelpout Type Iii Antifreeze Protein A20L Mutant (Nfe6, Afp), P21 Form
Organism: Zoarces elongatus
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2018-05-16 Classification: ANTIFREEZE PROTEIN |
 |
Crystal Structure Of Notched-Fin Eelpout Type Iii Antifreeze Protein A20T Mutant (Nfe6, Afp), P21 Form
Organism: Zoarces elongatus
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2018-05-16 Classification: ANTIFREEZE PROTEIN |
 |
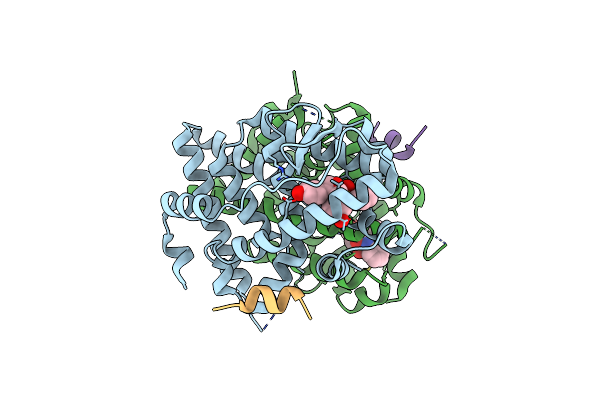
Crystal Structure Of Rhodopsin Bound To Visual Arrestin Determined By X-Ray Free Electron Laser
Organism: Enterobacteria phage rb55, Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2017-08-09 Classification: SIGNALING PROTEIN |
 |
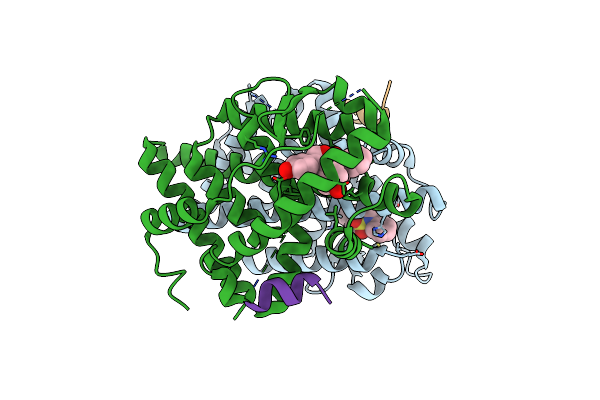
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (Y537S) In Complex With Oxabicyclic Heptene Sulfonamide (Obhs-N)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2016-11-16 Classification: TRANSCRIPTION Ligands: OB1 |
 |
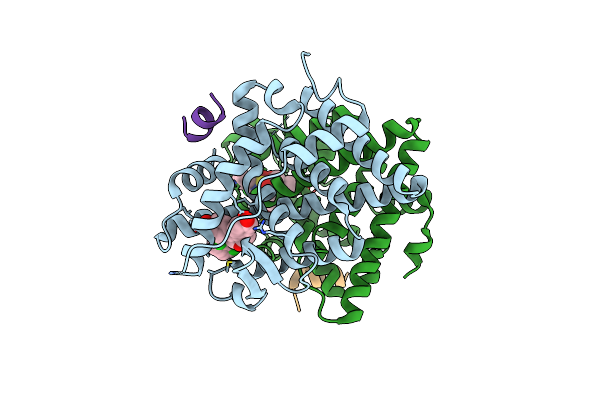
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (Y537S) In Complex With An N-Methyl Substituted Obhs-N Derivative
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2016-11-16 Classification: TRANSCRIPTION Ligands: OB2 |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (Y537S) In Complex With An N-Methyl, 2-Chlorobenzyl Obhs-N Derivative
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-11-16 Classification: TRANSCRIPTION Ligands: OB3 |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (Y537S) In Complex With An N-Ethyl, 4-Methoxybenzyl Obhs-N Derivative
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2016-11-16 Classification: TRANSCRIPTION Ligands: OB5, OB4 |

