Search Count: 28
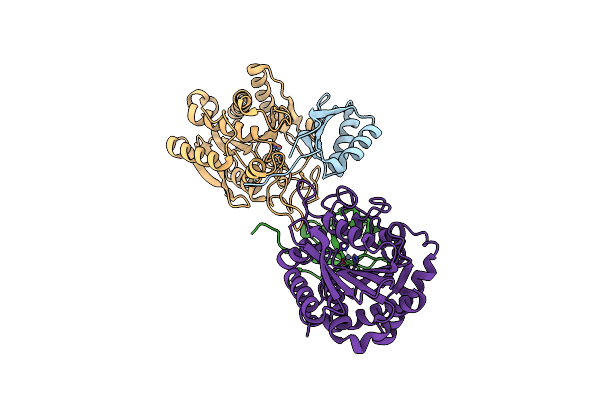 |
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2021-11-10 Classification: HYDROLASE Ligands: ZN |
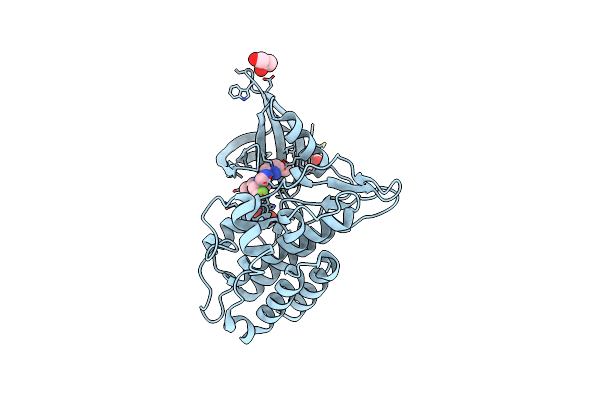 |
Tgf-Beta Receptor Type 1 Kinase Domain (T204D) In Complex With 6-[1-(2,2-Difluoroethyl)-4-(6-Methylpyridin-2-Yl)-1H-Imidazol-5-Yl]Imidazo[1,2-A]Pyridine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2020-02-05 Classification: TRANSFERASE Ligands: QMY, GOL |
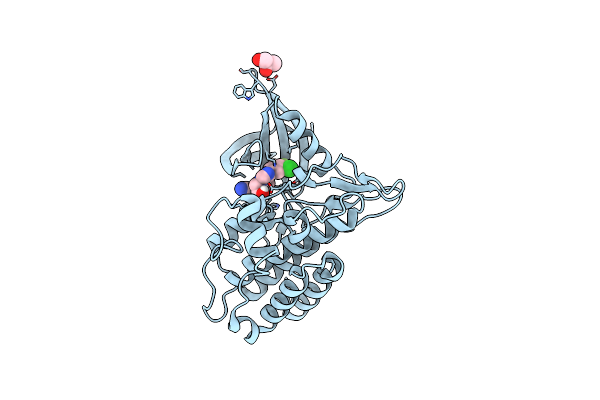 |
Tgf-Beta Receptor Type 1 Kinase Domain (T204D) In Complex With 6-[4-(3-Chloro-4-Fluorophenyl)-1-(2-Hydroxyethyl)-1H-Imidazol-5-Yl]Imidazo[1,2-B]Pyridazine-3-Carbonitrile
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2020-02-05 Classification: TRANSFERASE Ligands: QMV, GOL |
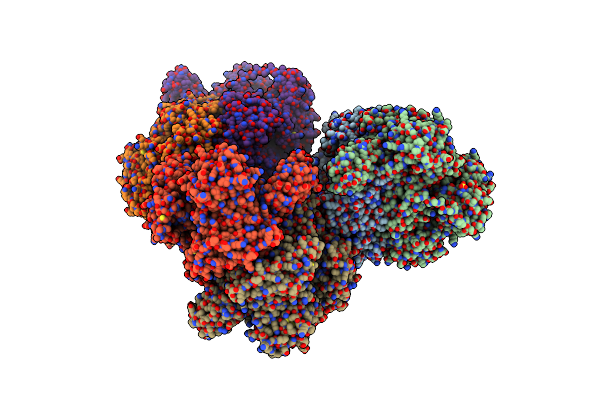 |
Crystal Structure Of Pyruvate:Ferredoxin Oxidoreductase From Moorella Thermoacetica
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-03-28 Classification: OXIDOREDUCTASE Ligands: SF4, TPP, MG, SO4 |
 |
Pyruvate:Ferredoxin Oxidoreductase From Moorella Thermoacetica With Lactyl-Tpp Bound
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-03-28 Classification: OXIDOREDUCTASE Ligands: SF4, TDL, MG, SO4, TPP |
 |
Pyruvate:Ferredoxin Oxidoreductase From Moorella Thermoacetica With Acetyl-Tpp Bound
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2018-03-28 Classification: OXIDOREDUCTASE Ligands: SF4, HTL, MG, SO4 |
 |
Pyruvate:Ferredoxin Oxidoreductase From Moorella Thermoacetica With Coenzyme A Bound
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2018-03-28 Classification: OXIDOREDUCTASE Ligands: SF4, TPP, MG, COA |
 |
Tgf-Beta Receptor Type 1 Kinase Domain (T204D) In Complex With N-(3-Fluoropyridin-4-Yl)-2-[6-(Trifluoromethyl)Pyridin-2-Yl]-7H-Pyrrolo[2,3-D]Pyrimidin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-02-07 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: D0A |
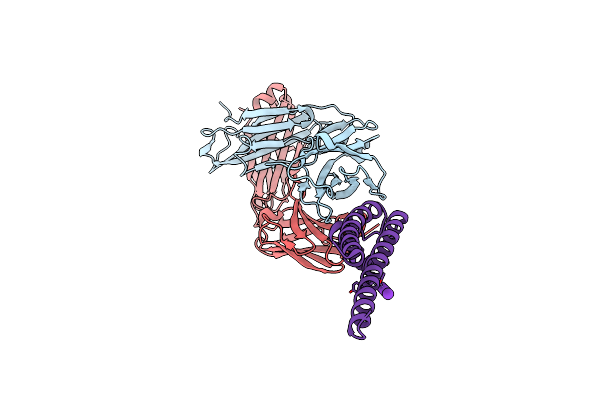 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-04-18 Classification: MEMBRANE PROTEIN Ligands: K, CD |
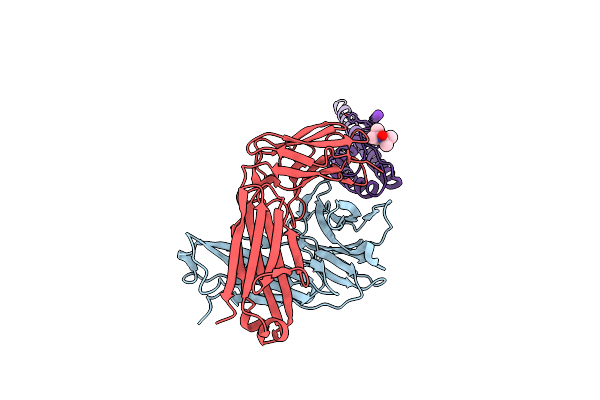 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-04-18 Classification: MEMBRANE PROTEIN Ligands: MTN, K |
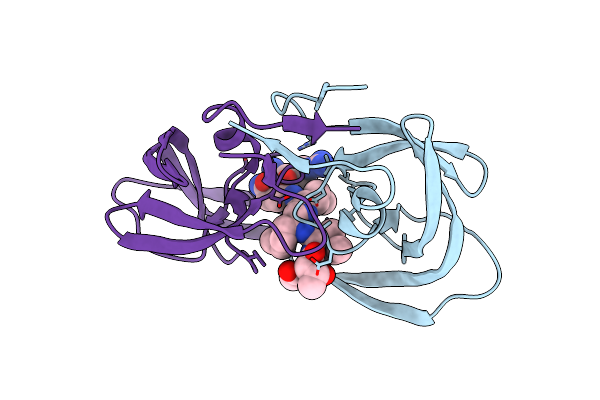 |
X-Ray Structure Of Ester Chemical Analogue [O-Ile50,O-Ile50']Hiv-1 Protease Complexed With Mvt-101
Method: X-RAY DIFFRACTION
Resolution:1.50 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: 2NC |
 |
X-Ray Structure Of Ester Chemical Analogue [O-Ile50,O-Ile50']Hiv-1 Protease Complexed With Kvs-1 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.90 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: SO4, KVS |
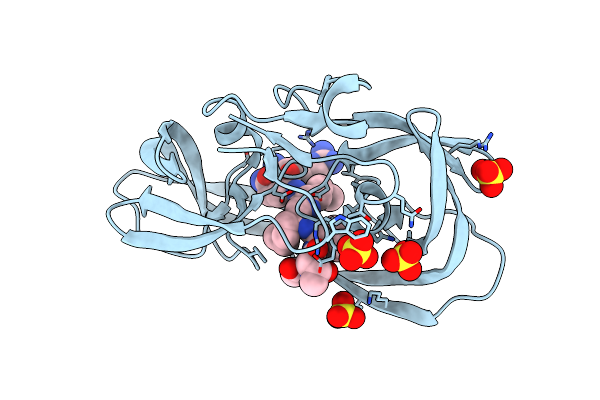 |
X-Ray Structure Of Ester Chemical Analogue 'Covalent Dimer' [Ile50,O-Ile50']Hiv-1 Protease Complexed With Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.61 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: 2NC, SO4 |
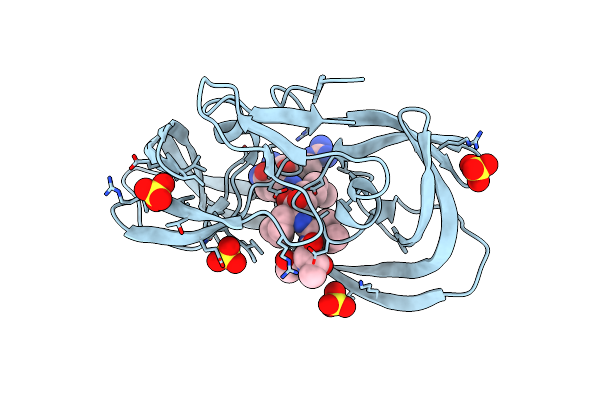 |
X-Ray Structure Of Ester Chemical Analogue 'Covalent Dimer' [Ile50,O-Ile50']Hiv-1 Protease Complexed With Kvs-1 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.80 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: KVS, SO4 |
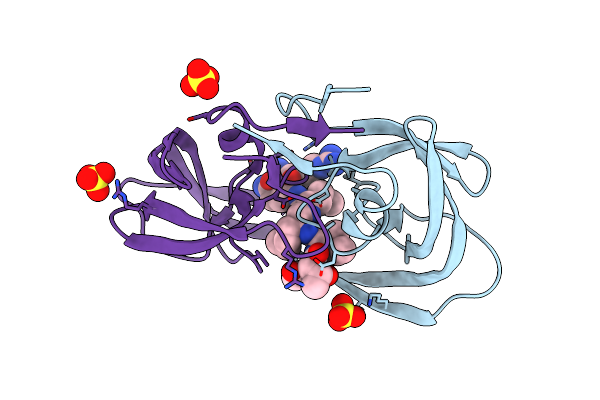 |
X-Ray Structure Of Ester Chemical Analogue [O-Gly51,O-Gly51']Hiv-1 Protease Complexed With Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.45 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: 2NC, SO4 |
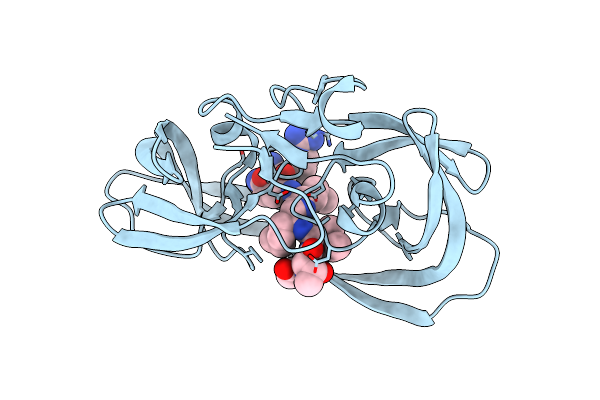 |
Crystal Structure Of Chemically Synthesized 'Covalent Dimer' [Gly51/D-Ala51']Hiv-1 Protease
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2011-07-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |
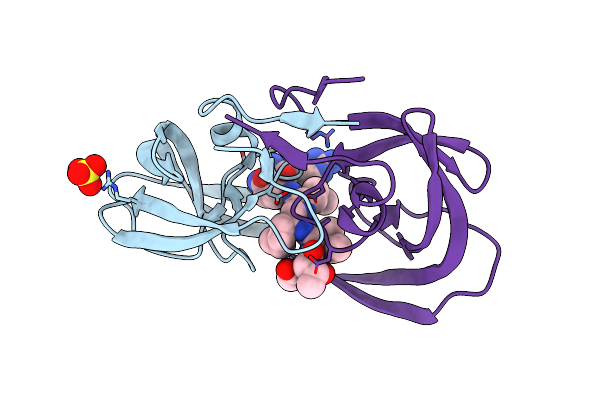 |
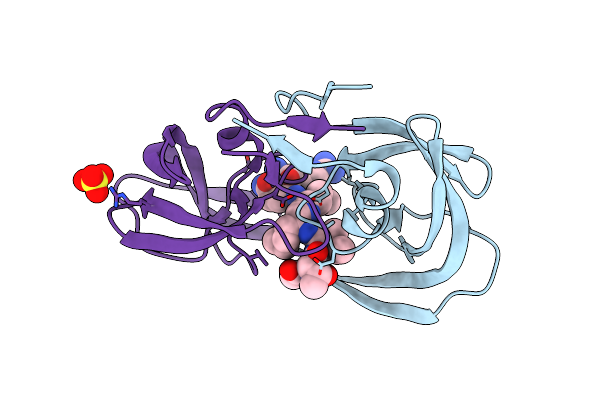
Crystal Structure Of Chemically Synthesized Hiv-1 Protease With Reduced Isostere Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.30 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
 |
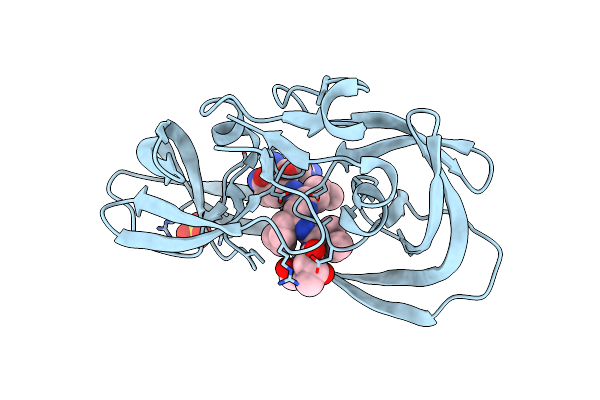
Crystal Structure Of [L-Ala51/51']Hiv-1 Protease With Reduced Isostere Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.30 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
 |
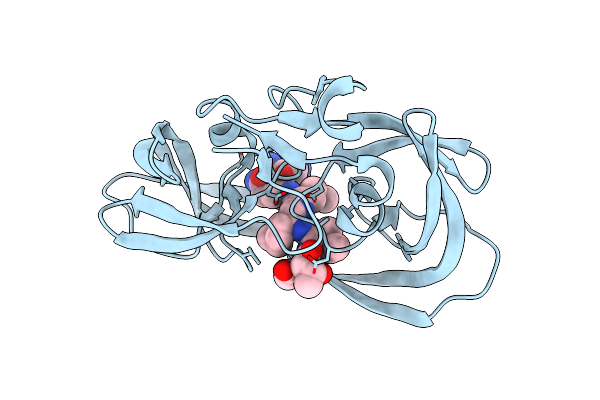
Crystal Structure Of A Chemically Synthesized 203 Amino Acid 'Covalent Dimer' [Gly51;Aib51']Hiv-1 Protease Molecule Complexed With Mvt-101 Reduced Isostere Inhibitor At 1.6 A Resolution
Method: X-RAY DIFFRACTION
Resolution:1.61 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
 |
Crystal Structure Of Chemically Synthesized 203 Amino Acid 'Covalent Dimer' [L-Ala;Gly51']Hiv-1 Protease Molecule Complexed With Mvt-101 Reduced Isostere Inhibitor At 1.4 A Resolution
Method: X-RAY DIFFRACTION
Resolution:1.40 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |

