Search Count: 12
 |
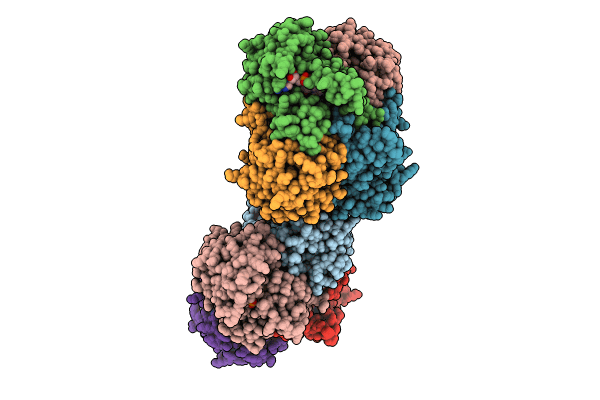
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: NAD, DMS, PEG, PO4 |
 |
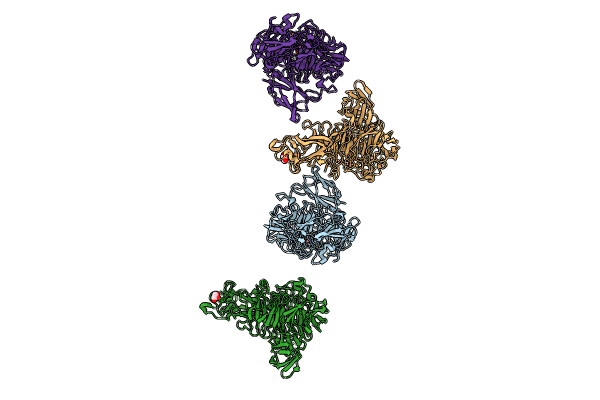
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: NAD, CL |
 |
Crystal Structure Of An Engineered Variant Of Galactose Oxidase, Goaserd4Bb, From Fusarium Graminearum
Organism: Fusarium graminearum
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: CU, GOL |
 |
Crystal Structure Of An Engineered Variant Of Galactose Oxidase, Goaserd7Bb, From Fusarium Graminearum
Organism: Fusarium graminearum
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: ACT, CU, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2022-04-06 Classification: IMMUNE SYSTEM Ligands: ZEV |
 |
Crystal Structure Of An Engineered Variant Of Single-Chain Penicillin G Acylase From Kluyvera Cryocrescens (Global Hydrolysis Rd3Chis)
Organism: Kluyvera cryocrescens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2021-11-17 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of An Engineered Variant Of Single-Chain Penicillin G Acylase From Kluyvera Cryocrescens (A1-Ac Rd3Chis)
Organism: Kluyvera cryocrescens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-11-17 Classification: HYDROLASE Ligands: CA, PMS |
 |
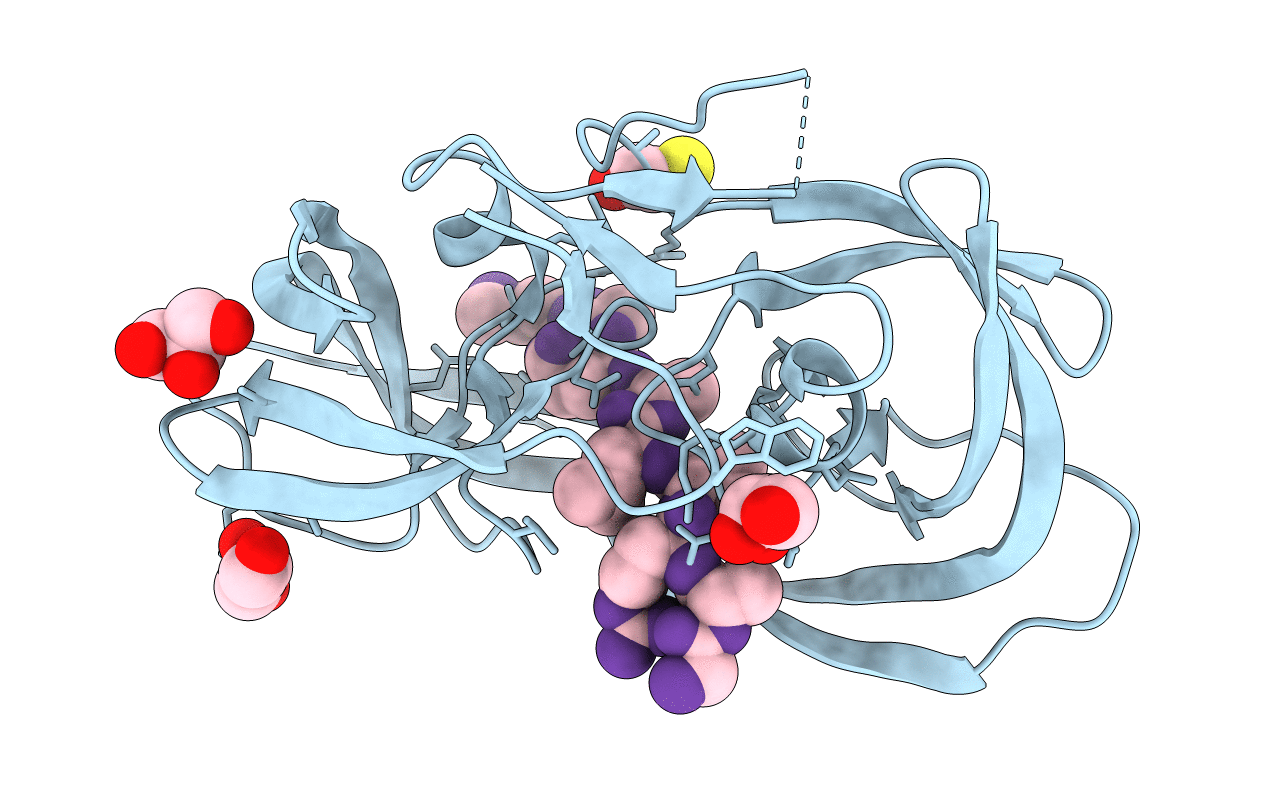
Crystal Structure Of Inactive Single Chain Wild-Type Hiv-1 Protease In Complex With The Substrate Rt-Rh
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: GOL, PO4 |
 |
Crystal Structure Of Inactive Single Chain Wild-Type Hiv-1 Protease In Complex With The Substrate Ca-P2
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: GOL, BME |
 |
Crystal Structure Of Inactive Single Chain Wild-Type Hiv-1 Protease In Complex With The Substrate P2-Nc
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: GOL, EDO, DMS, ACT, BME |
 |
Crystal Structure Of Inactive Single Chain Variant Of Hiv-1 Protease In Complex With The Substrate P2-Nc
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: BME, EDO, GOL, ACT, PO4 |
 |
Crystal Structure Of Inactive Single Chain Variant Of Hiv-1 Protease In Complex With The Substrate Rt-Rh
Organism: Hiv-1 m:b_arv2/sf2, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-06-06 Classification: hydrolase/hydrolase substrate Ligands: EDO, GOL |

