Search Count: 12
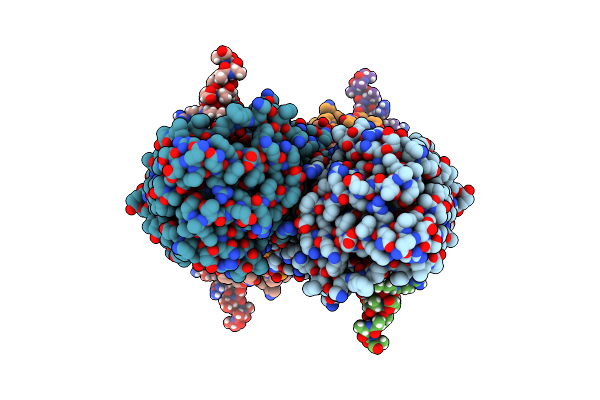 |
Partial Structure Of Tyrosine Hydroxylase Lacking The First 35 Residues In Complex With Dopamine.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: OXIDOREDUCTASE Ligands: LDP, FE |
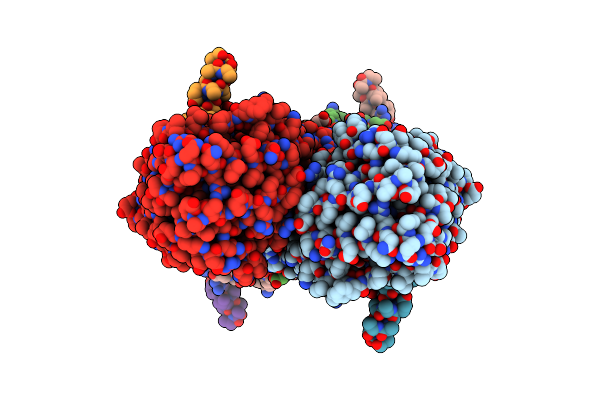 |
Partial Structure Of Tyrosine Hydroxylase In Complex With Dopamine Showing The Catalytic Domain And An Alpha-Helix From The Regulatory Domain Involved In Dopamine Binding.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-08 Classification: OXIDOREDUCTASE Ligands: LDP, FE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: OXIDOREDUCTASE Ligands: FE |
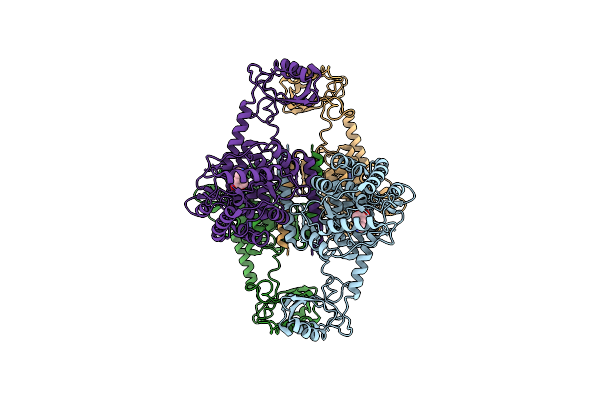 |
Atomic Model Of The Em-Based Structure Of The Full-Length Tyrosine Hydroxylase In Complex With Dopamine (Residues 40-497) In Which The Regulatory Domain (Residues 40-165) Has Been Included Only With The Backbone Atoms
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-17 Classification: OXIDOREDUCTASE Ligands: FE, LDP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-17 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-25 Classification: PLANT PROTEIN |
 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-12-25 Classification: PLANT PROTEIN |
 |
Crystal Structure Of The Carboxy-Terminal Region Of The Bacteriophage T4 Proximal Long Tail Fibre Protein Gp34, Residues 795 To 1289, At 1.9 Angstrom.
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-07-12 Classification: VIRAL PROTEIN Ligands: GOL, PO4, ACT, NA, URE |
 |
Crystal Structure Of The Carboxy-Terminal Region Of The Bacteriophage T4 Proximal Long Tail Fibre Protein Gp34, Residues 744-1289 At 2.9 Angstrom Resolution
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2017-07-12 Classification: VIRAL PROTEIN Ligands: GOL |
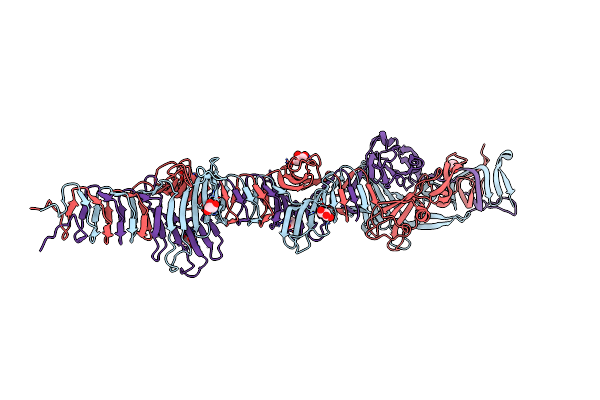 |
Crystal Structure Of The Carboxy-Terminal Region Of The Bacteriophage T4 Proximal Long Tail Fibre Protein Gp34, P21 Native Crystal
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-12-16 Classification: VIRAL PROTEIN Ligands: GOL |
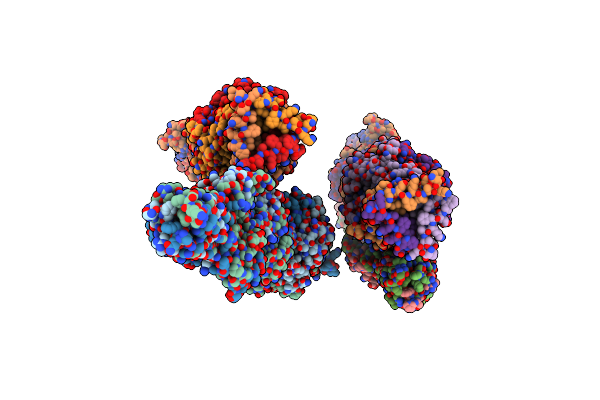 |
Crystal Structure Of The Carboxy-Terminal Region Of The Bacteriophage T4 Proximal Long Tail Fibre Protein Gp34, R32 Native Crystal
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-09-30 Classification: VIRAL PROTEIN |
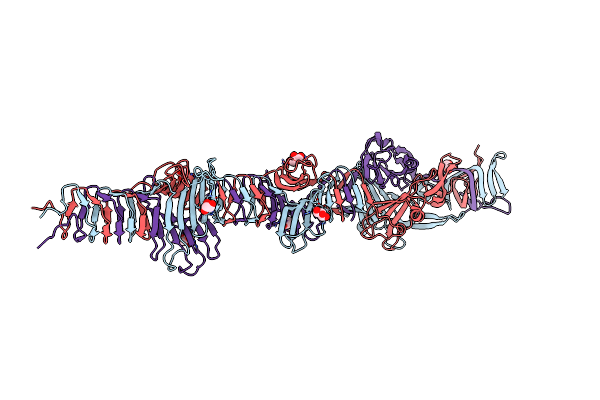 |
Crystal Structure Of The Carboxy-Terminal Region Of The Bacteriophage T4 Proximal Long Tail Fibre Protein Gp34, P21 Selenomethionine Crystal
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-08-19 Classification: VIRAL PROTEIN Ligands: GOL |

