Search Count: 36
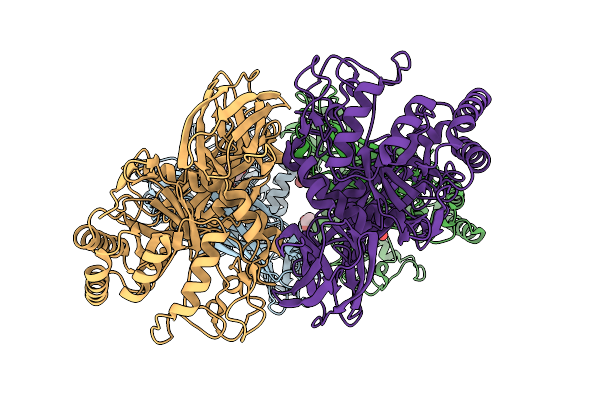 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum, Wild Type Semet Derivative (Lagh157)
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: ACT, GOL, MLA |
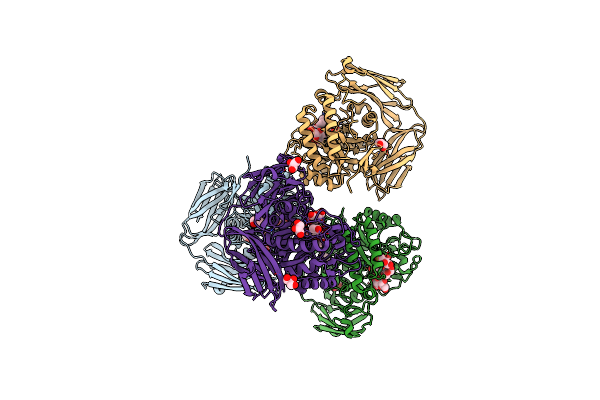 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum (Lagh157) E224A Mutant In Complex With Laminaritriose And Glucose
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: EDO, BGC, GOL, PGE |
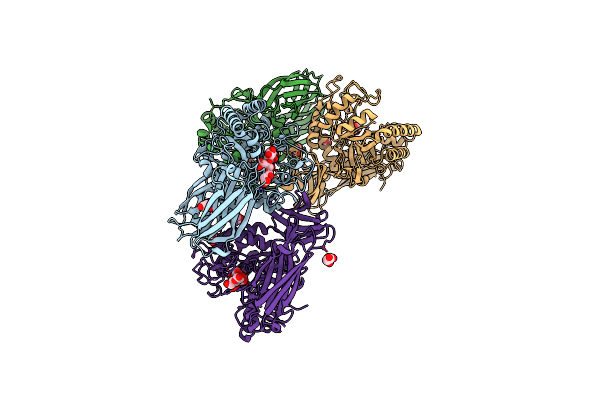 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum (Lagh157) In Complex With Laminaribiose
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: MLA, GOL, BGC, PG4 |
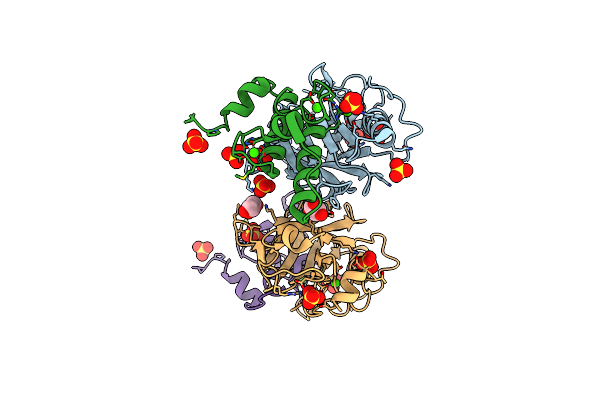 |
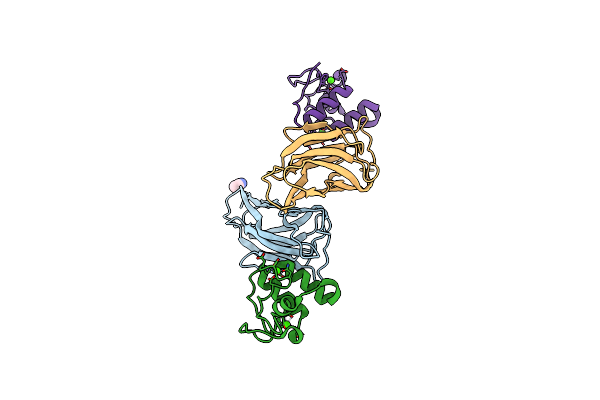
Ruminococcus Flavefaciens Cohesin-Dockerin Structure: Dockerin From Scah Adaptor Scaffoldin In Complex With The Cohesin From Scae Anchoring Scaffoldin
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-11-02 Classification: PROTEIN BINDING Ligands: CA, GOL, SO4 |
 |
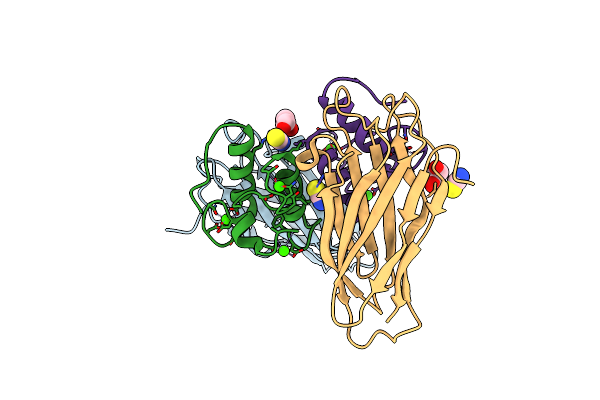
Crystal Structure Of Ruminococcus Flavefaciens' Type Iii Complex Containing The Fifth Cohesin From Scaffoldin B And The Dockerin From Scaffoldin A
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-02-28 Classification: PROTEIN BINDING Ligands: CCN, CA |
 |
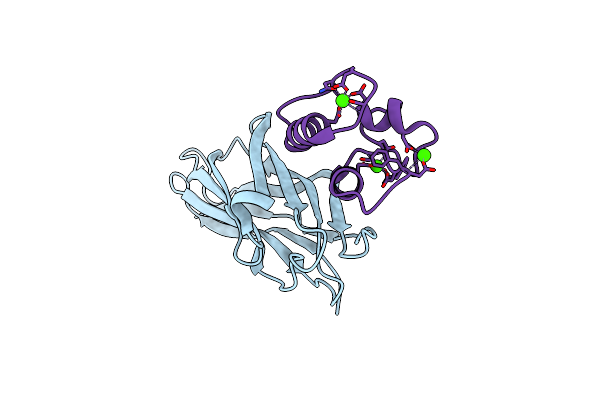
Crystal Structure Of The Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B In Complex With Cel5 Dockerin S15I, I16N Mutant
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-01-31 Classification: PROTEIN BINDING Ligands: CA, SCN, GOL |
 |
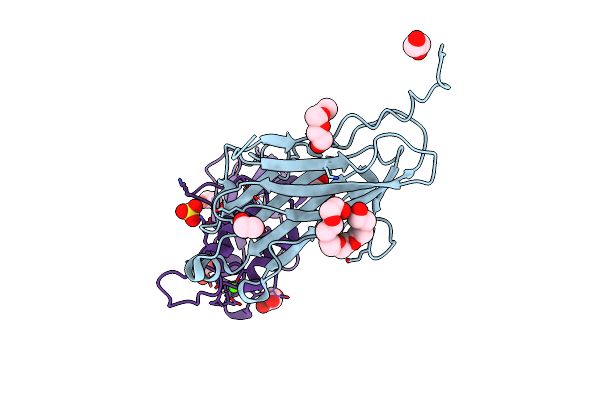
Crystal Structure Of The Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B In Complex With Cel5 Dockerin S51I, L52N Mutant
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-01-31 Classification: CELL ADHESION Ligands: CA |
 |
Crystal Structure Of The Cohscaa-Xdoccipb Type Ii Complex From Clostridium Thermocellum At 1.5Angstrom Resolution
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-09-06 Classification: CELL ADHESION Ligands: CA, SO4, GOL, EDO, PGE, P6G |
 |
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2017-07-05 Classification: PROTEIN BINDING Ligands: CA |
 |
Organism: Ruminococcus flavefaciens, Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-07-05 Classification: PROTEIN BINDING Ligands: CA, GOL |
 |
Crystal Structure Of The Cohscac2-Xdoccipa Type Ii Complex From Clostridium Thermocellum
Organism: Ruminiclostridium thermocellum ad2, Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-04-05 Classification: CARBOHYDRATE BINDING PROTEIN Ligands: CA |
 |
Organism: Ruminiclostridium thermocellum 27405, Clostridium thermocellum 27405
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-03-29 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
Organism: Spirochaeta thermophila (strain atcc 700085 / dsm 6578 / z-1203)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-02-15 Classification: SUGAR BINDING PROTEIN Ligands: 2NV, SSN, CA, NI, FMT |
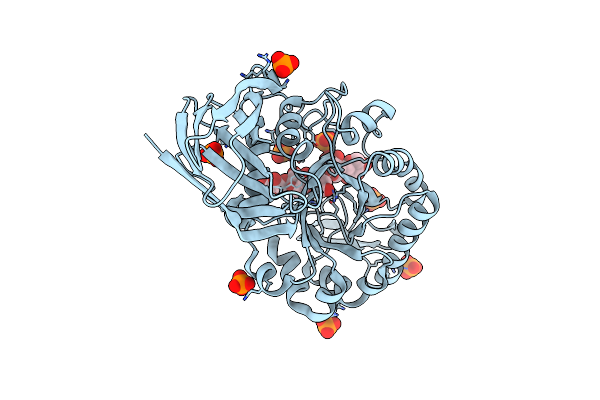 |
High Resolution Structure Of The Thermostable Glucuronoxylan Endo-Beta-1, 4-Xylanase, Ctxyn30A, From Clostridium Thermocellum With Two Xylobiose Units Bound
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-19 Classification: HYDROLASE Ligands: PO4, PEG |
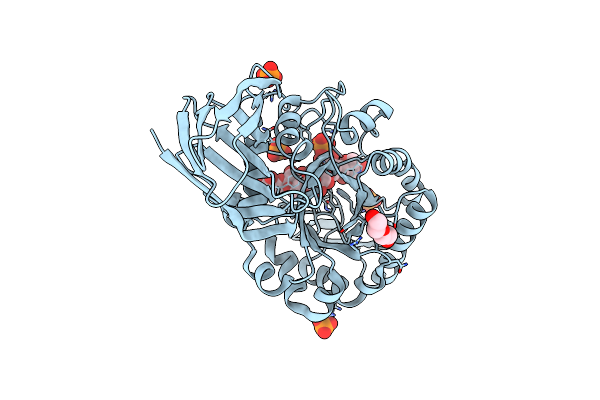 |
Determining The Specificities Of The Catalytic Site From The Very High Resolution Structure Of The Thermostable Glucuronoxylan Endo-Beta-1, 4-Xylanase, Ctxyn30A, From Clostridium Thermocellum With A Xylotetraose Bound
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2016-10-19 Classification: HYDROLASE Ligands: PO4, PEG |
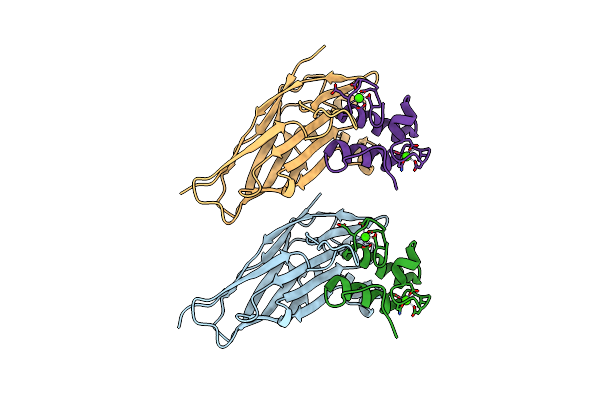 |
Crystal Structure Of Ruminococcus Flavefaciens Scaffoldin C Cohesin In Complex With A Dockerin From An Uncharacterized Cbm-Containing Protein
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-10-19 Classification: PROTEIN BINDING Ligands: CA |
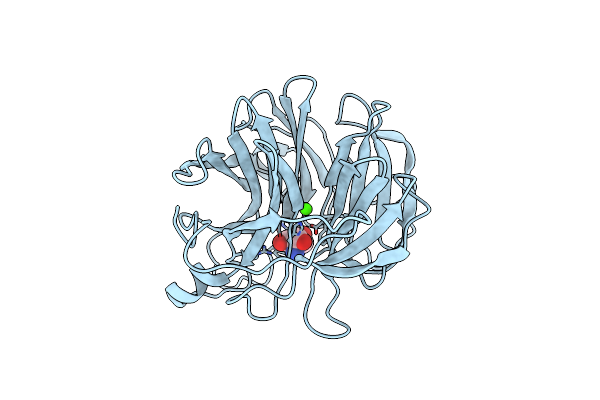 |
The Ultra High Resolution Structure Of A Novel Alpha-L-Arabinofuranosidase (Ctgh43) From Clostridium Thermocellum Atcc 27405 With Bound Trimethyl N-Oxide (Trs)
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2016-07-27 Classification: HYDROLASE Ligands: CA, TRS |
 |
The High Resolution Structure Of A Novel Alpha-L-Arabinofuranosidase (Ctgh43) From Clostridium Thermocellum Atcc 27405
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-07-27 Classification: HYDROLASE Ligands: SO4, ACT, GOL |
 |
Crystal Structure Of Coh3Scab-Xdoc_M2Scaa Complex: A C-Terminal Interface Mutant Of Type Ii Cohesin-X-Dockerin Complex From Acetivibrio Cellulolyticus
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2015-10-07 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
Crystal Structure Of Coh3Scab-Xdoc_M1Scaa Complex: A N-Terminal Interface Mutant Of Type Ii Cohesin-X-Dockerin Complex From Acetivibrio Cellulolyticus
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2015-07-29 Classification: PROTEIN BINDING Ligands: NHE, CA |

