Search Count: 27
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2021-04-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: Y5Y, EDO, SO4 |
 |
Tfe-Induced Nmr Structure Of A Novel Bioactive Peptide (Padbs1R2) Derived From A Pyrobaculum Aerophilum Ribosomal Protein (L39E)
Organism: Pyrobaculum aerophilum
Method: SOLUTION NMR Release Date: 2019-09-25 Classification: ANTIMICROBIAL PROTEIN |
 |
Tfe-Induced Nmr Structure Of A Novel Bioactive Peptide (Padbs1R3) Derived From A Pyrobaculum Aerophilum Ribosomal Protein (L39E)
Organism: Pyrobaculum aerophilum
Method: SOLUTION NMR Release Date: 2019-09-25 Classification: ANTIMICROBIAL PROTEIN |
 |
Tfe-Induced Nmr Structure Of A Novel Bioactive Peptide (Padbs1R7) Derived From A Pyrobaculum Aerophilum Ribosomal Protein (L39E)
Organism: Pyrobaculum aerophilum
Method: SOLUTION NMR Release Date: 2019-09-25 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2019-05-01 Classification: TRANSFERASE Ligands: K4A |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2019-05-01 Classification: TRANSFERASE Ligands: K47 |
 |
3D Structure Of E. Coli Isocitrate Dehydrogenase K100M Mutant In Complex With Isocitrate And Magnesium(Ii)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-05-21 Classification: OXIDOREDUCTASE Ligands: ICT, MG, SO4 |
 |
3D Structure Of E. Coli Isocitrate Dehydrogenase In Complex With Isocitrate, Calcium(Ii) And Nadp - The Pseudo-Michaelis Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-10-31 Classification: OXIDOREDUCTASE Ligands: NAP, ICT, CA |
 |
3D Structure Of E. Coli Isocitrate Dehydrogenase In Complex With Isocitrate, Calcium(Ii) And Thionadp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-10-31 Classification: OXIDOREDUCTASE Ligands: TAP, ICT, CA, SO4 |
 |
3D Structure Of E. Coli Isocitrate Dehydrogenase K100M Mutant In Complex With Isocitrate, Magnesium(Ii) And Thionadp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-10-31 Classification: OXIDOREDUCTASE Ligands: TAP, ICT, MG, SO4 |
 |
3D Structure Of E. Coli Isocitrate Dehydrogenase K100M Mutant In Complex With Alpha-Ketoglutarate, Calcium(Ii) And Adenine Nucleotide Phosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-10-31 Classification: OXIDOREDUCTASE Ligands: A2P, AKG, CA, SO4 |
 |
3D Structure Of E. Coli Isocitrate Dehydrogenase K100M Mutant In Complex With Alpha-Ketoglutarate, Magnesium(Ii) And Nadph - The Product Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2012-10-31 Classification: OXIDOREDUCTASE Ligands: NMN, NAP, AKG, MG |
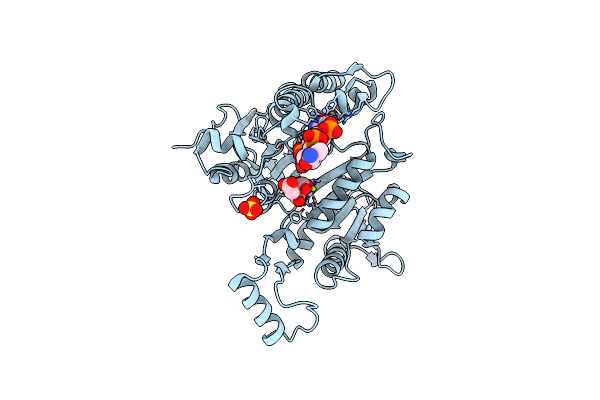 |
3D Structure Of E. Coli Isocitrate Dehydrogenase K100M Mutant In Complex With Isocitrate, Magnesium(Ii), Adenosine 2',5'-Biphosphate And Ribosylnicotinamide-5'-Phosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-10-31 Classification: OXIDOREDUCTASE Ligands: A2P, ICT, MG, NMN, SO4 |
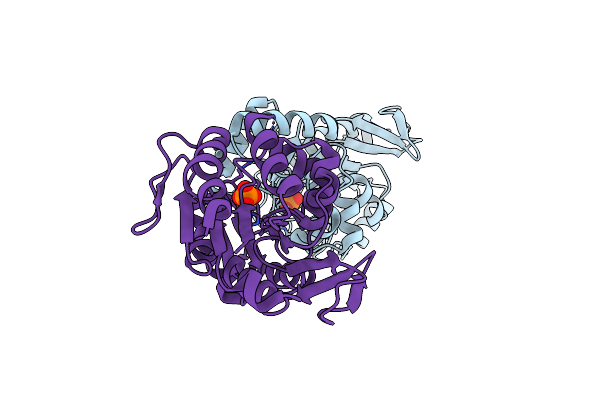 |
The 3-Dimensional Structure Of Apo-Mpgp, The Mannosyl-3- Phosphoglycerate Phosphatase From Thermus Thermophilus Hb27 In Its Apo-Form
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: PO4 |
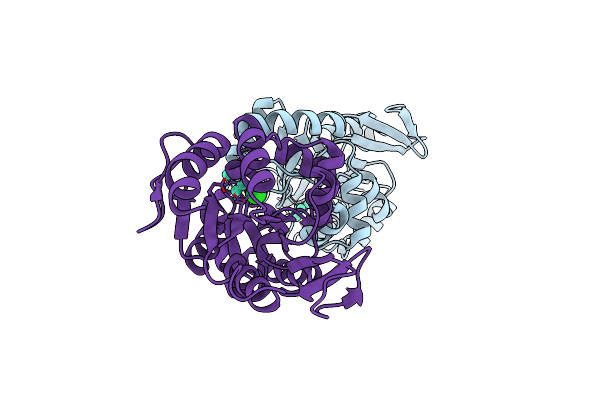 |
The 3-Dimensional Structure Of The Gadolinium Derivative Of Mpgp, The Mannosyl-3-Phosphoglycerate Phosphatase From Thermus Thermophilus Hb27
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: GD, CL |
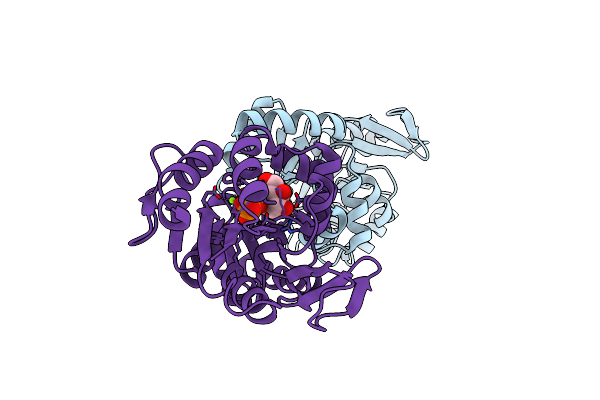 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Alpha-Mannosylglycerate And Orthophosphate Reaction Products.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: MG, PO4, 2M8 |
 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Alpha-Mannosylglycerate And Orthophosphate Reaction Products.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: MG, PO4, 2M8 |
 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Alpha-Mannosylglycerate And Metaphosphate.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: MG, PO4, CL, PO3, 2M8 |
 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Alpha-Mannosylglycerate.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: 2M8, MG |
 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Metavanadate
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: MG, VN3 |

