Search Count: 7
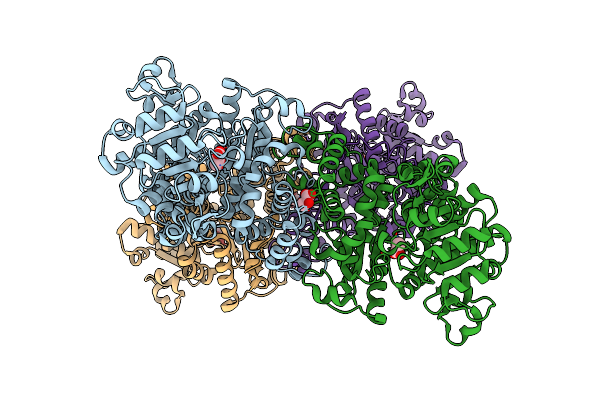 |
Crystal Structure Of The G200R Mutant From The Maize Chloroplastic Photosynthetic Nadp(+)-Dependent Malic Enzyme
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-01-15 Classification: PHOTOSYNTHESIS Ligands: PYR |
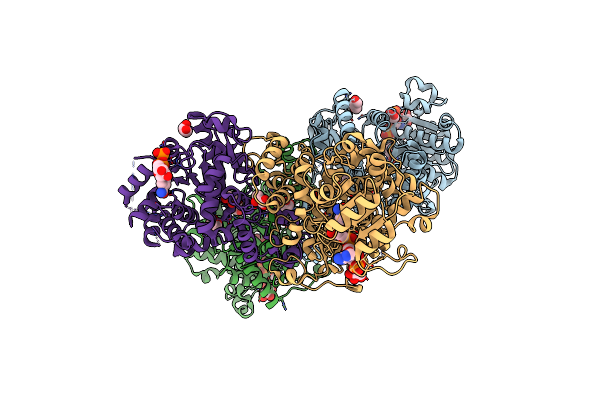 |
Organism: Sorghum bicolor
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-30 Classification: OXIDOREDUCTASE Ligands: NAP, GOL, PYR, PEG |
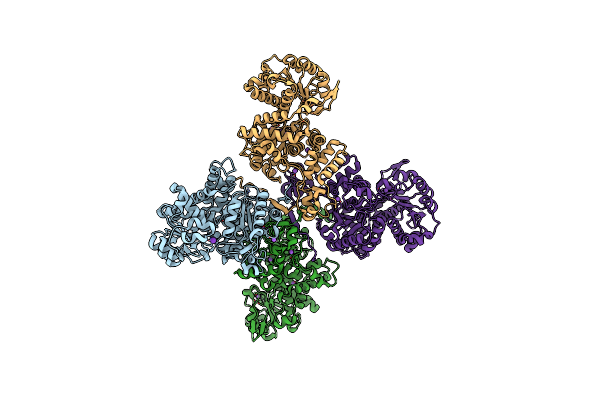 |
Crystal Structure Of Maize Chloroplastic Photosynthetic Nadp(+)-Dependent Malic Enzyme
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-12-12 Classification: PHOTOSYNTHESIS |
 |
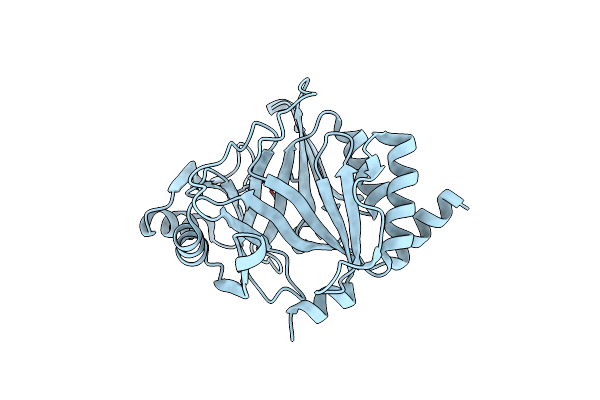
Crystal Structure Of V278E-Glyoxalase I Mutant From Zea Mays In Space Group P4(1)2(1)2
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-11-21 Classification: PLANT PROTEIN Ligands: GSH, CO, FMT |
 |
Crystal Structure Of V278E-Glyoxalase I Mutant From Zea Mays In Space Group P6(3)
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-11-21 Classification: PLANT PROTEIN Ligands: CO |
 |
Crystal Structure Of E144Q-Glyoxalase I Mutant From Zea Mays In Space Group P4(1)2(1)2
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-11-21 Classification: PLANT PROTEIN Ligands: CO, GSH, FMT |
 |
Organism: Aster yellows witches'-broom phytoplasma (strain aywb)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-10-05 Classification: OXIDOREDUCTASE Ligands: NAD, MG |

