Search Count: 17
 |
Organism: Escherichia coli w
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-05 Classification: TRANSCRIPTION Ligands: GOL, SO4 |
 |
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2021-10-06 Classification: PEPTIDE BINDING PROTEIN Ligands: PLP, GOL, CL |
 |
Organism: Staphylococcus aureus (strain mu50 / atcc 700699)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2014-09-10 Classification: HYDROLASE Ligands: CD, CL, MUR |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2014-05-21 Classification: HYDROLASE Ligands: CD, CL |
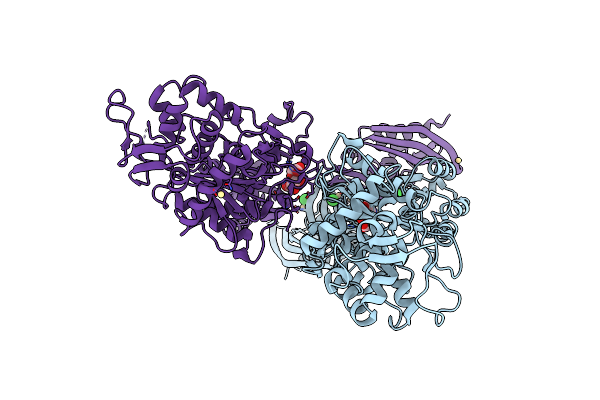 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-05-21 Classification: HYDROLASE Ligands: CD, CL, MUR |
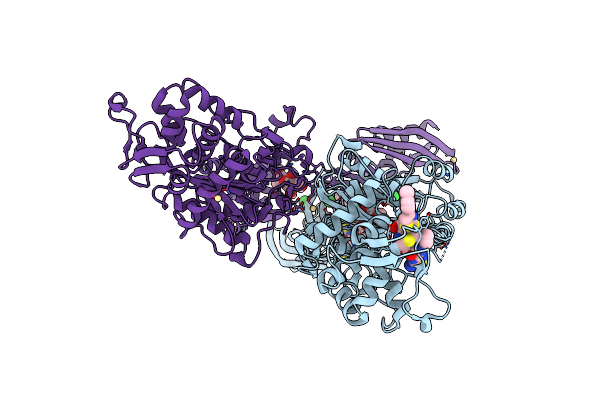 |
Crystal Structure Of Ceftaroline Acyl-Pbp2A From Mrsa With Non- Covalently Bound Ceftaroline And Muramic Acid At Allosteric Site Obtained By Soaking
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-10-09 Classification: HYDROLASE Ligands: CD, CL, AI8, 1W8, MUR |
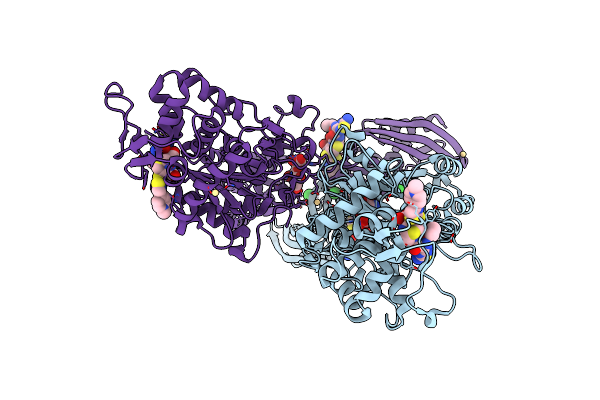 |
Crystal Structure Of Ceftaroline Acyl-Pbp2A From Mrsa With Non- Covalently Bound Ceftaroline And Muramic Acid At Allosteric Site Obtained By Cocrystallization
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-10-09 Classification: HYDROLASE Ligands: AI8, CD, CL, 1W8, MUR |
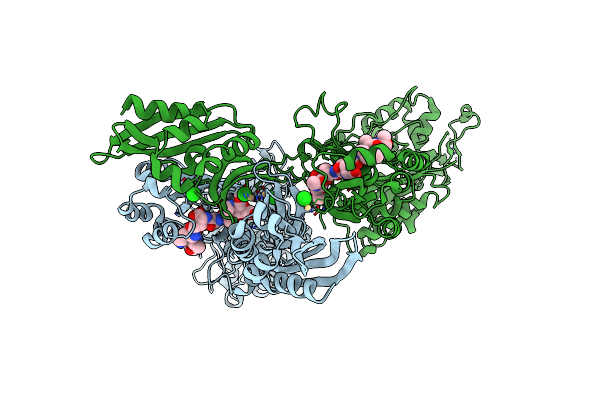 |
Crystal Structure Of Pbp2A From Mrsa In Complex With Peptidoglycan Analogue At Allosteric
Organism: Staphylococcus aureus subsp. aureus mu50, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2013-10-09 Classification: HYDROLASE Ligands: CD, CL, AMV |
 |
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-07-20 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-07-20 Classification: HYDROLASE Ligands: AH0, MHI, ZN |
 |
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-07-20 Classification: HYDROLASE |
 |
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-07-20 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-07-20 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2011-05-25 Classification: ELECTRON TRANSPORT Ligands: HEM, MG, SO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-04-13 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-11-20 Classification: ELECTRON TRANSPORT Ligands: MG, HEM |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-09-19 Classification: ELECTRON TRANSPORT Ligands: MG, HEM |

