Search Count: 42
 |
Impacts Of Ribosomal Rna Sequence Variation On Gene Expression And Phenotype: Cryo-Em Structure Of The Rrsb Ribosome (Bbb-70S)
Organism: Escherichia coli k-12, Escherichia coli bl21
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RIBOSOME Ligands: PUT, SPD, MG, K, ZN, ATP, FME |
 |
Impacts Of Ribosomal Rna Sequence Variation On Gene Expression And Phenotype: Cryo-Em Structure Of The Rrsh Ribosome (Hbb-70S)
Organism: Escherichia coli k-12, Escherichia coli bl21
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RIBOSOME Ligands: PUT, SPD, MG, K, ZN, ATP, FME |
 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Ic State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, PUT, K, MG, ANM, 3H3, ZN, MET |
 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Ga State)
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, K, MG, ANM, PUT, 3H3, ZN, PHE, MET, GSP, ZIY |
 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Cr State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, PUT, MG, ANM, 3H3, K, ZN, PHE, MET, GSP, ZIY |
 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Ac State)
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: PUT, K, MG, ANM, SPD, 3H3, ZN, MET |
 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Ga State 2)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, K, MG, PUT, 3HE, HMT, ZN, PHE, MET, GSP, YRB |
 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Consensus Lsu Focused Refined Structure)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, PUT, K, MG, ANM, 3H3, TRS, ZN, MET |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-03-16 Classification: DE NOVO PROTEIN |
 |
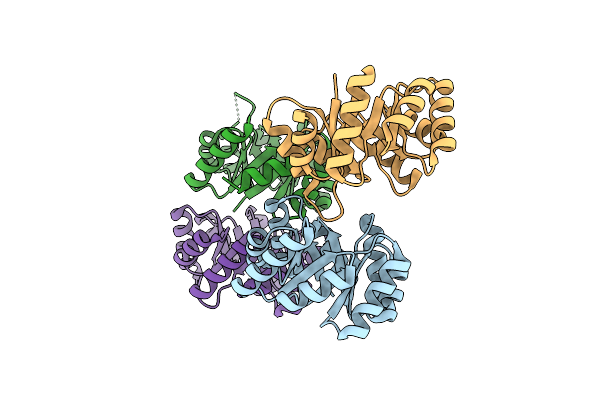
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-01-19 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-01-19 Classification: DE NOVO PROTEIN |
 |
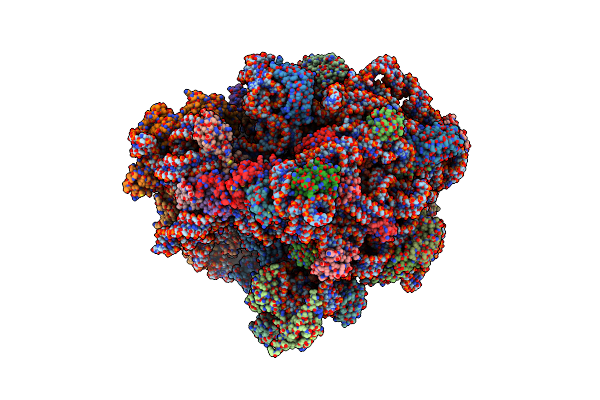
Elongating 70S Ribosome Complex In A Classical Pre-Translocation (Pre-C) Conformation
Organism: Escherichia coli k-12, Escherichia coli bl21, Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: RIBOSOME Ligands: PUT, MG, ZN, ATP, SPD |
 |
Elongating 70S Ribosome Complex In A Fusidic Acid-Stalled Intermediate State Of Translocation Bound To Ef-G(Gdp) (Int2)
Organism: Escherichia coli (strain k12), Escherichia coli, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: RIBOSOME Ligands: PUT, MG, ZN, ATP, SPD, FUA, GDP |
 |
Elongating 70S Ribosome Complex In A Hybrid-H1 Pre-Translocation (Pre-H1) Conformation
Organism: Escherichia coli k-12, Escherichia coli bl21, Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: RIBOSOME Ligands: PUT, MG, ZN, ATP, SPD |
 |
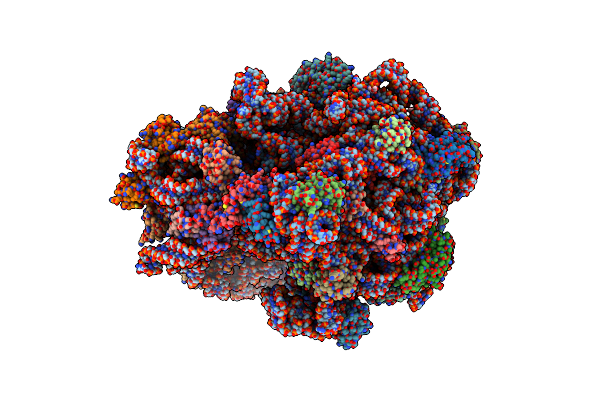
Elongating 70S Ribosome Complex In A Spectinomycin-Stalled Intermediate State Of Translocation Bound To Ef-G In An Active, Gtp Conformation (Int1)
Organism: Escherichia coli k-12, Escherichia coli bl21
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: RIBOSOME Ligands: SCM, PUT, MG, ZN, ATP, SPD, GTP |
 |
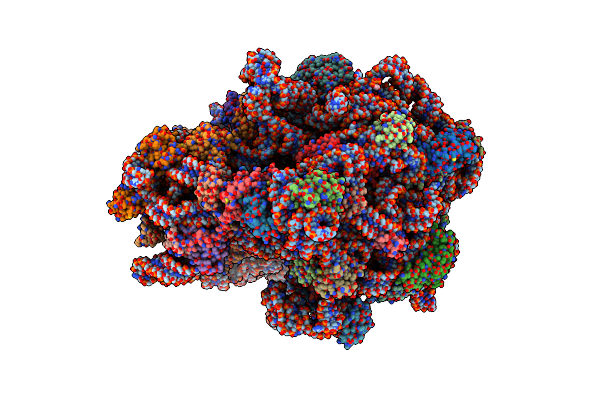
Elongating 70S Ribosome Complex In A Hybrid-H2* Pre-Translocation (Pre-H2*) Conformation
Organism: Escherichia coli k-12, Escherichia coli bl21, Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: RIBOSOME Ligands: PUT, MG, ZN, ATP, SPD |
 |
Elongating 70S Ribosome Complex In A Post-Translocation (Post) Conformation
Organism: Escherichia coli k-12, Escherichia coli bl21
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: RIBOSOME Ligands: MG, ATP, PUT, SPD, ZN |
 |
Crystal Structure Of Aminoglycoside Tc007 In Complex With 70S Ribosome From Thermus Thermophilus, Three Trnas And Mrna (Soaking)
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2018-05-02 Classification: RIBOSOME Ligands: 8UZ, MG, ZN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2017-12-13 Classification: RIBOSOME Ligands: MG, GET, ZN |
 |
Crystal Structure Of Aminoglycoside Tc007 Co-Crystallized With 70S Ribosome From Thermus Thermophilus, Three Trnas And Mrna
Organism: Escherichia coli, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2017-12-13 Classification: RIBOSOME Ligands: 8UZ, MG, ZN |

