Search Count: 8
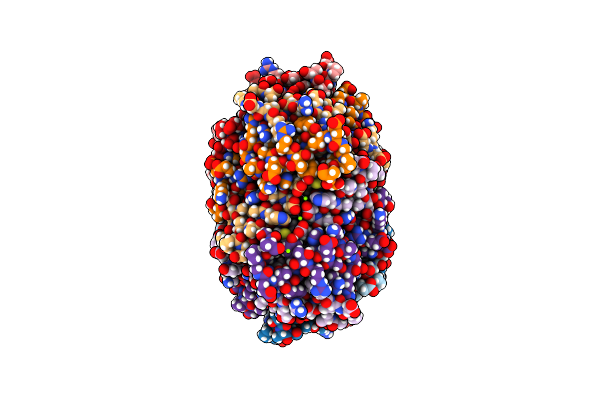 |
Crystal Structure Of Rhodospirillum Rubrum Rru_A0973 Mutant E32A, E62A, H65A.
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: MG, PGE |
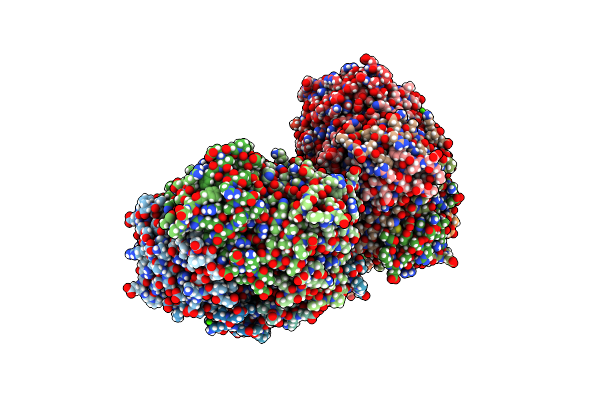 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: CA |
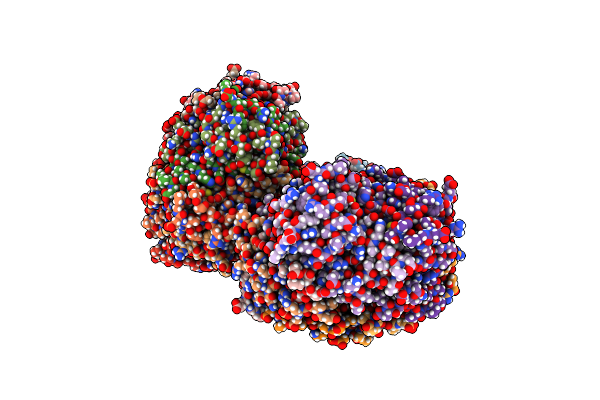 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: CA |
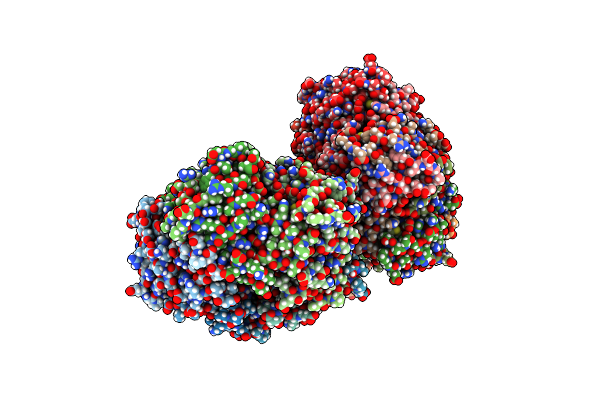 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2016-08-10 Classification: OXIDOREDUCTASE Ligands: FE, GOA, CA |
 |
Structure Of A C269A Mutant Of Propionaldehyde Dehydrogenase From The Clostridium Phytofermentans Fucose Utilisation Bacterial Microcompartment
Organism: Clostridium phytofermentans (strain atcc 700394 / dsm 18823 / isdg)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2016-03-16 Classification: OXIDOREDUCTASE Ligands: SO4, COA, ACT |
 |
Structure Of His387Ala Mutant Of The Propionaldehyde Dehydrogenase From The Clostridium Phytofermentans Fucose Utilisation Bacterial Microcompartment
Organism: Clostridium phytofermentans
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2016-03-16 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Structure Of A Propionaldehyde Dehydrogenase From The Clostridium Phytofermentans Fucose Utilisation Bacterial Microcompartment
Organism: Clostridium phytofermentans
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2013-09-11 Classification: OXIDOREDUCTASE Ligands: SO4, NAD |

