Search Count: 19
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, ACT, GOL |
 |
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, SO4, EDO |
 |
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG |
 |
Crystal Structure Of Lotus Japonicus Chip13 Extracellular Domain In Complex With A Nanobody
Organism: Lotus japonicus, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG |
 |
Crystal Structure Of Lotus Japonicus Chip13 Extracellular Domain In Complex With Chitooctaose
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: GOL, NAG |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN |
 |
Organism: Lotus japonicus, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, EDO |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: ACT, IMD, GOL, SO4, NAG |
 |
Crystal Structure Of Lotus Japonicus Chip13 Extracellular Domain In Complex With Chitooctaose
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: IMD, EDO |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, EDO, MAN |
 |
Organism: Mycobacteroides abscessus atcc 19977
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-05-10 Classification: UNKNOWN FUNCTION Ligands: PO4, GOL |
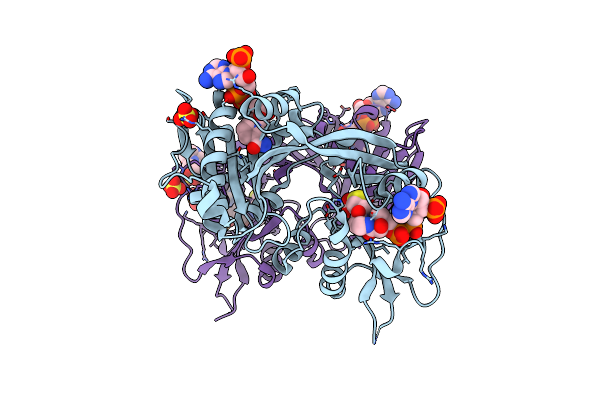 |
Crystal Structure Of Mab_4324 A Tandem Repeat Gnat From Mycobacterium Abscessus
Organism: Mycobacteroides abscessus (strain atcc 19977 / dsm 44196 / cip 104536 / jcm 13569 / nctc 13031 / tmc 1543)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: NDP, COA, ACO, ACT, SO4, NAI, EPE |
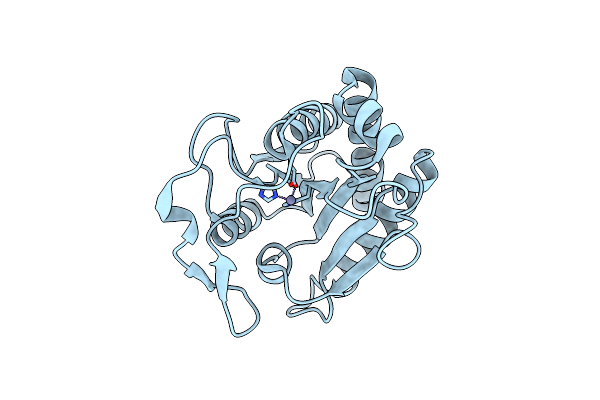 |
Crystal Structure Of The Apo Form Of The N-Acetylmuramyl-L-Alanine Amidase, Ami1, From Mycobacterium Abscessus.
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-18 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
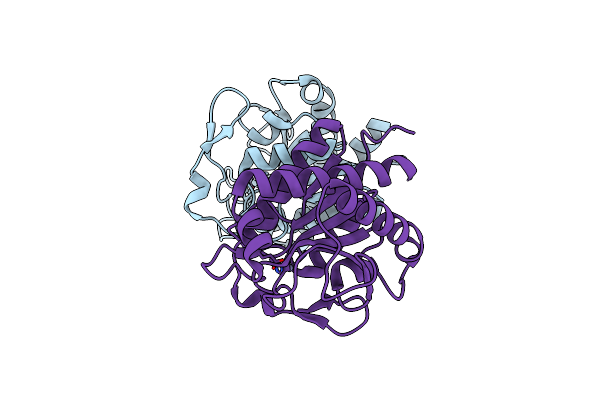 |
Crystal Structure Of The N-Acetylmuramyl-L-Alanine Amidase, Ami1, From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-11-18 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
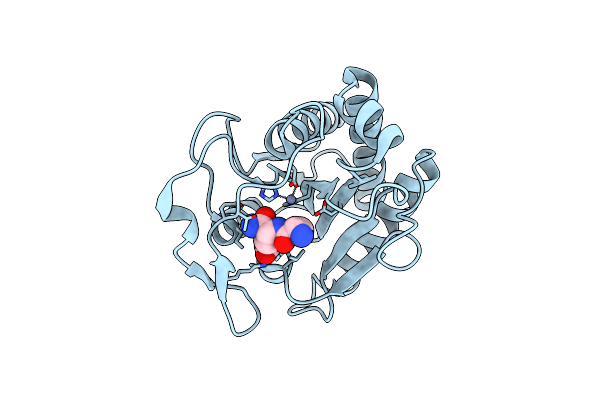 |
Crystal Structure Of The N-Acetylmuramyl-L-Alanine Amidase, Ami1, From Mycobacterium Abscessus Bound To L-Alanine-D-Isoglutamine
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-11-18 Classification: SUGAR BINDING PROTEIN Ligands: ZGL, ALA, ZN |
 |
Crystal Structure Of The Mycobacterial Trehalose Monomycolate Transport Factor A, Ttfa
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-12-25 Classification: LIPID TRANSPORT Ligands: SO4 |
 |
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-10 Classification: ANTIBIOTIC Ligands: ACO |
 |
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-07-10 Classification: ANTIBIOTIC Ligands: CIT, P4K, ACT |
 |
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-07-10 Classification: ANTIBIOTIC Ligands: BTB, SO4 |

